1VDR
 
 | | DIHYDROFOLATE REDUCTASE | | Descriptor: | DIHYDROFOLATE REDUCTASE, PHOSPHATE ION | | Authors: | Pieper, U, Herzberg, O. | | Deposit date: | 1997-11-30 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural features of halophilicity derived from the crystal structure of dihydrofolate reductase from the Dead Sea halophilic archaeon, Haloferax volcanii.
Structure, 6, 1998
|
|
1S2T
 
 | | Crystal Structure Of Apo Phosphoenolpyruvate Mutase | | Descriptor: | Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1S2U
 
 | | Crystal structure of the D58A phosphoenolpyruvate mutase mutant protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1RXX
 
 | | Structure of arginine deiminase | | Descriptor: | Arginine deiminase | | Authors: | Galkin, A, Kulakova, L, Sarikaya, E, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insight into arginine degradation by arginine deiminase, an antibacterial and parasite drug target.
J.Biol.Chem., 279, 2004
|
|
1S2V
 
 | | Crystal structure of phosphoenolpyruvate mutase complexed with Mg(II) | | Descriptor: | MAGNESIUM ION, Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1GGO
 
 | | T453A MUTANT OF PYRUVATE, PHOSPHATE DIKINASE | | Descriptor: | PROTEIN (PYRUVATE, PHOSPHATE DIKINASE), SULFATE ION | | Authors: | Li, Z, Herzberg, O. | | Deposit date: | 2000-08-29 | | Release date: | 2001-01-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of domain-domain docking sites within Clostridium symbiosum pyruvate phosphate dikinase by amino acid replacement.
J.Biol.Chem., 275, 2000
|
|
7RFO
 
 | | SeMet Tailspike protein 4 (TSP4) phage CBA120, residues 1-335, obtained in the presence of LiSO4 | | Descriptor: | Tailspike protein | | Authors: | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | Deposit date: | 2021-07-14 | | Release date: | 2022-02-23 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
7REJ
 
 | | Tailspike protein 4 (TSP4) from phage CBA120, residues 1-335, obtained in the presence of NaK-Tartrate | | Descriptor: | IMIDAZOLE, Tailspike protein | | Authors: | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | Deposit date: | 2021-07-13 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
7RFV
 
 | | Tailspike protein 4 (TSP4) from phage CBA120, residues 1-250, obtained in the presence of PEG8000 | | Descriptor: | Tailspike protein | | Authors: | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | Deposit date: | 2021-07-14 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
7PWO
 
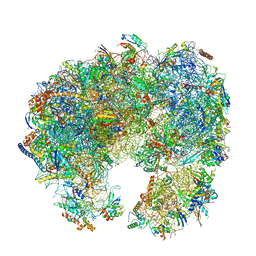 | | Cryo-EM structure of Giardia lamblia ribosome at 2.75 A resolution | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S26, 40S ribosomal protein S30, ... | | Authors: | Hiregange, D.G, Rivalta, A, Bose, T, Breiner-Goldstein, E, Samiya, S, Cimicata, G, Kulakova, L, Zimmerman, E, Bashan, A, Herzberg, O, Yonath, A. | | Deposit date: | 2021-10-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Cryo-EM structure of the ancient eukaryotic ribosome from the human parasite Giardia lamblia.
Nucleic Acids Res., 50, 2022
|
|
7PWG
 
 | | Cryo-EM structure of large subunit of Giardia lamblia ribosome at 2.7 A resolution | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L18a, 60S ribosomal protein L27, ... | | Authors: | Hiregange, D.G, Rivalta, A, Bose, T, Breiner-Goldstein, E, Samiya, S, Cimicata, G, Kulakova, L, Zimmerman, E, Bashan, A, Herzberg, O, Yonath, A. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Cryo-EM structure of the ancient eukaryotic ribosome from the human parasite Giardia lamblia.
Nucleic Acids Res., 50, 2022
|
|
7PWF
 
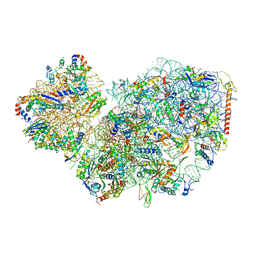 | | Cryo-EM structure of small subunit of Giardia lamblia ribosome at 2.9 A resolution | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | Authors: | Hiregange, D.G, Rivalta, A, Bose, T, Breiner-Goldstein, E, Samiya, S, Cimicata, G, Kulakova, L, Zimmerman, E, Bashan, A, Herzberg, O, Yonath, A. | | Deposit date: | 2021-10-06 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of the ancient eukaryotic ribosome from the human parasite Giardia lamblia.
Nucleic Acids Res., 50, 2022
|
|
2OUT
 
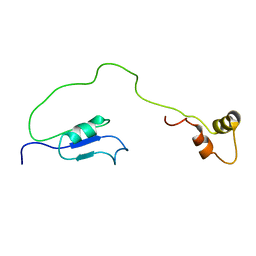 | | Solution Structure of HI1506, a Novel Two Domain Protein from Haemophilus influenzae | | Descriptor: | Mu-like prophage FluMu protein gp35, Protein HI1507 in Mu-like prophage FluMu region | | Authors: | Sari, N, He, Y, Doseeva, V, Surabian, K, Schwarz, F, Herzberg, O, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2007-02-12 | | Release date: | 2007-05-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HI1506, a novel two-domain protein from Haemophilus influenzae.
Protein Sci., 16, 2007
|
|
2PE4
 
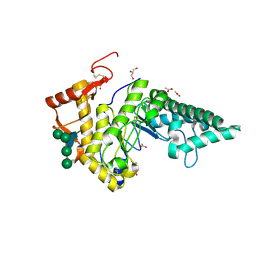 | | Structure of Human Hyaluronidase 1, a Hyaluronan Hydrolyzing Enzyme Involved in Tumor Growth and Angiogenesis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Chao, K.L, Herzberg, O. | | Deposit date: | 2007-04-02 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Human Hyaluronidase-1, a Hyaluronan Hydrolyzing Enzyme Involved in Tumor Growth and Angiogenesis
Biochemistry, 46, 2007
|
|
4OJ6
 
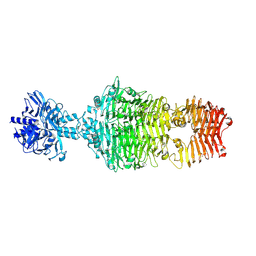 | |
4OJ5
 
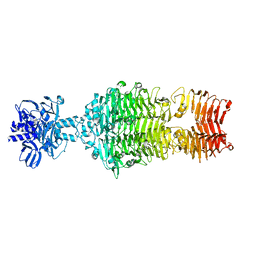 | |
8P33
 
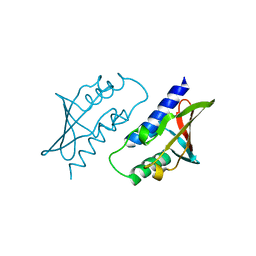 | | BB0238 from Borrelia burgdorferi | | Descriptor: | BB0238 | | Authors: | Brangulis, K, Foor, S.D, Shakya, A.K, Rana, V.S, Bista, S, Kitsou, C, Ronzetti, M, Linden, S.B, Altieri, A.S, Akopjana, I, Baljinnyam, B, Nelson, D.C, Simeonov, A, Herzberg, O, Caimano, M.J, Pal, U. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A unique borrelial protein facilitates microbial immune evasion.
Mbio, 14, 2023
|
|
3QYN
 
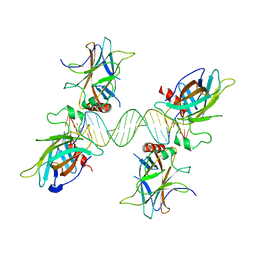 | |
8P32
 
 | | BB0238 from Borrelia burgdorferi, Se-Met data for Leu240Met mutant | | Descriptor: | BB0238 | | Authors: | Brangulis, K, Foor, S.D, Shakya, A.K, Rana, V.S, Bista, S, Kitsou, C, Ronzetti, M, Linden, S.B, Altieri, A.S, Akopjana, I, Baljinnyam, B, Nelson, D.C, Simeonov, A, Herzberg, O, Caimano, M.J, Pal, U. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A unique borrelial protein facilitates microbial immune evasion.
Mbio, 14, 2023
|
|
3US1
 
 | |
3US2
 
 | |
3US0
 
 | |
5TIB
 
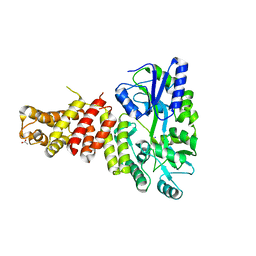 | | Gasdermin-B C-terminal domain containing the polymorphism residues Arg299:Ser306 fused to maltose binding protein | | Descriptor: | ACETATE ION, SODIUM ION, Sugar ABC transporter substrate-binding protein,Gasdermin-B, ... | | Authors: | Chao, K, Herzberg, O. | | Deposit date: | 2016-10-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human Gasdermin-B and disease: Sulfatide Binding, Caspase cleavage, and Structural impact of Asthma- and IBS-Associated Polymorphism
Proc.Natl.Acad.Sci.Usa, 2017
|
|
3KZF
 
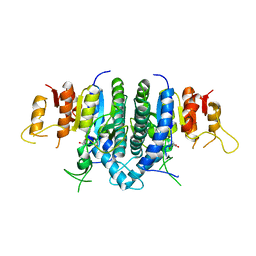 | | Structure of Giardia Carbamate Kinase | | Descriptor: | Carbamate kinase, GLYCEROL | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2009-12-08 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure and characterization of carbamate kinase from the human parasite Giardia lamblia.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5TJ4
 
 | |
