7D9B
 
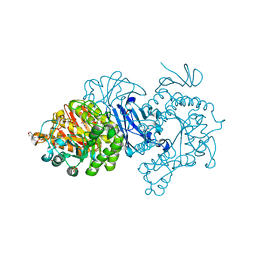 | | Crystal structure of alpha-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-glycosidase, CALCIUM ION, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-12 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DCG
 
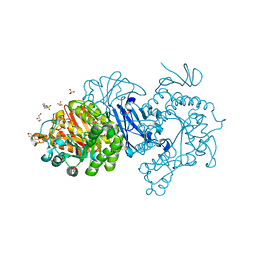 | | Alpha-glucosidase from Weissella cibaria BBK-1 bound with maltotriose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-glycosidase, CALCIUM ION, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1UIX
 
 | | Coiled-coil structure of the RhoA-binding domain in Rho-kinase | | Descriptor: | Rho-associated kinase | | Authors: | Shimizu, T, Ihara, K, Maesaki, R, Amano, M, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2003-07-23 | | Release date: | 2003-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parallel coiled-coil association of the RhoA-binding domain in Rho-kinase
J.Biol.Chem., 278, 2003
|
|
7EVT
 
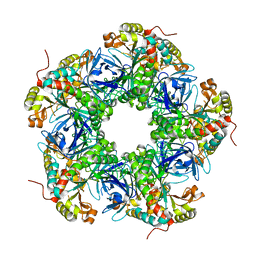 | | Crystal structure of the N-terminal degron-truncated human glutamine synthetase | | Descriptor: | Glutamine synthetase | | Authors: | Chek, M.F, Kim, S.Y, Mori, T, Hakoshima, T. | | Deposit date: | 2021-05-22 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of N-terminal degron-truncated human glutamine synthetase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
1IS7
 
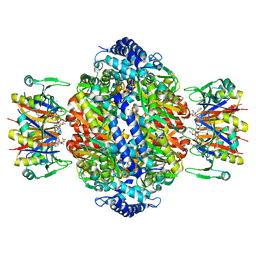 | | Crystal structure of rat GTPCHI/GFRP stimulatory complex | | Descriptor: | GTP Cyclohydrolase I, GTP Cyclohydrolase I Feedback Regulatory Protein, PHENYLALANINE, ... | | Authors: | Maita, N, Okada, K, Hatakeyama, K, Hakoshima, T. | | Deposit date: | 2001-11-18 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the stimulatory complex of GTP cyclohydrolase I and its feedback regulatory protein GFRP.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7EHI
 
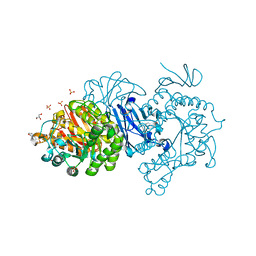 | | Crystal structure of covalent maltosyl-alpha-glucosidase intermediate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EHH
 
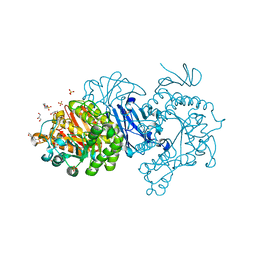 | | Crystal structure of alpha-glucosidase from Weissella cibaria BKK1 in complex with maltose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1DJ6
 
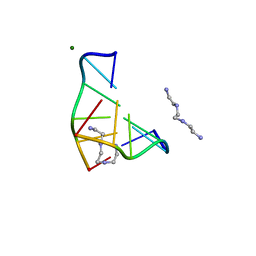 | | COMPLEX OF A Z-DNA HEXAMER, D(CG)3, WITH SYNTHETIC POLYAMINE AT ROOM TEMPERATURE | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', MAGNESIUM ION, N,N'-BIS(2-AMINOETHYL)-1,2-ETHANEDIAMINE | | Authors: | Ohishi, H, Tomita, K.-i, Nakanishi, I, Ohtsuchi, M, Hakoshima, T, Rich, A. | | Deposit date: | 1999-12-01 | | Release date: | 1999-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The crystal structure of N1-[2-(2-amino-ethylamino)-ethyl]-ethane-1,2-diamine (polyamines) binding to the minor groove of d(CGCGCG)2, hexamer at room temperature
FEBS Lett., 523, 2002
|
|
3WX2
 
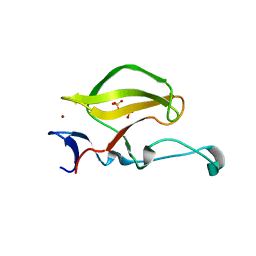 | | Mouse Cereblon thalidomide binding domain, native | | Descriptor: | Protein cereblon, SULFATE ION, ZINC ION | | Authors: | Mori, T, Ito, T, Hirano, Y, Yamaguchi, Y, Handa, H, Hakoshima, T. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs
Nat.Struct.Mol.Biol., 21, 2014
|
|
3WX1
 
 | | Mouse Cereblon thalidomide binding domain, selenomethionine derivative | | Descriptor: | Protein cereblon, SULFATE ION, ZINC ION | | Authors: | Mori, T, Ito, T, Hirano, Y, Yamaguchi, Y, Handa, H, Hakoshima, T. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs
Nat.Struct.Mol.Biol., 21, 2014
|
|
1UL1
 
 | | Crystal structure of the human FEN1-PCNA complex | | Descriptor: | Flap endonuclease-1, MAGNESIUM ION, Proliferating cell nuclear antigen | | Authors: | Sakurai, S, Kitano, K, Yamaguchi, H, Hamada, K, Okada, K, Fukuda, K, Uchida, M, Ohtsuka, E, Morioka, H, Hakoshima, T. | | Deposit date: | 2003-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for recruitment of human flap endonuclease 1 to PCNA
EMBO J., 24, 2005
|
|
1RLS
 
 | | CRYSTAL STRUCTURE OF RNASE T1 COMPLEXED WITH THE PRODUCT NUCLEOTIDE 3'-GMP. STRUCTURAL EVIDENCE FOR DIRECT INTERACTION OF HISTIDINE 40 AND GLUTAMIC ACID 58 WITH THE 2'-HYDROXYL GROUP OF RIBOSE | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Gohda, K, Oka, K.-I, Tomita, K.-I, Hakoshima, T. | | Deposit date: | 1994-03-29 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of RNase T1 complexed with the product nucleotide 3'-GMP. Structural evidence for direct interaction of histidine 40 and glutamic acid 58 with the 2'-hydroxyl group of the ribose.
J.Biol.Chem., 269, 1994
|
|
3W91
 
 | | crystal structure of SeMet-labeled yeast N-acetyltransferase Mpr1 L87M mutant | | Descriptor: | MPR1 protein | | Authors: | Nasuno, R, Hirano, Y, Itoh, T, Hakoshima, T, Hibi, T, Takagi, H. | | Deposit date: | 2013-03-23 | | Release date: | 2013-07-17 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional analysis of the yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3W6X
 
 | | Yeast N-acetyltransferase Mpr1 in complex with CHOP | | Descriptor: | (4S)-4-hydroxy-L-proline, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Nasuno, R, Hirano, Y, Itoh, T, Hakoshima, T, Hibi, T, Takagi, H. | | Deposit date: | 2013-02-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural and functional analysis of the yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3W6S
 
 | | yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, MPR1 protein | | Authors: | Nasuno, R, Hirano, Y, Itoh, T, Hakoshima, T, Hibi, T, Takagi, H. | | Deposit date: | 2013-02-21 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of the yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1VTG
 
 | | THE MOLECULAR STRUCTURE OF A DNA-TRIOSTIN A COMPLEX | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), triostin A | | Authors: | Wang, A.H.-J, Ughetto, G, Quigley, G.J, Hakoshima, T, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The molecular structure of a DNA-triostin A complex.
Science, 225, 1984
|
|
1VTW
 
 | | AT Base Pairs Are Less Stable than GC Base Pairs in Z-DNA: The Crystal Structure of D(M(5)CGTAM(5)CG) | | Descriptor: | DNA (5'-D(*(CH3)CP*GP*TP*AP*(CH3)CP*G)-3') | | Authors: | Wang, A.H.-J, Hakoshima, T, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | AT Base Pairs Are Less Stable than GC Base Pairs in Z-DNA: The Crystal Structure of d(m(5)CGTAm(5)CG)
Cell(Cambridge,Mass.), 37, 1984
|
|
3A8Q
 
 | | Low-resolution crystal structure of the Tiam2 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 2 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3A8N
 
 | | Crystal structure of the Tiam1 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 1 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3A8P
 
 | | Crystal structure of the Tiam2 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 2 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3W05
 
 | | Crystal structure of Oryza sativa DWARF14 (D14) in complex with PMSF | | Descriptor: | 1,2-ETHANEDIOL, Dwarf 88 esterase, phenylmethanesulfonic acid | | Authors: | Kagiyama, M, Hirano, Y, Mori, T, Kim, S.Y, Kyozuka, J, Seto, Y, Yamaguchi, S, Hakoshima, T. | | Deposit date: | 2012-10-19 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structures of D14 and D14L in the strigolactone and karrikin signaling pathways
Genes Cells, 18, 2013
|
|
3W06
 
 | | Crystal structure of Arabidopsis thaliana DWARF14 Like (AtD14L) | | Descriptor: | 1,2-ETHANEDIOL, Hydrolase, alpha/beta fold family protein | | Authors: | Kagiyama, M, Hirano, Y, Mori, T, Kim, S.Y, Kyozuka, J, Seto, Y, Yamaguchi, S, Hakoshima, T. | | Deposit date: | 2012-10-19 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structures of D14 and D14L in the strigolactone and karrikin signaling pathways
Genes Cells, 18, 2013
|
|
3W04
 
 | | Crystal structure of Oryza sativa DWARF14 (D14) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kagiyama, M, Hirano, Y, Mori, T, Kim, S.Y, Kyozuka, J, Seto, Y, Yamaguchi, S, Hakoshima, T. | | Deposit date: | 2012-10-19 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structures of D14 and D14L in the strigolactone and karrikin signaling pathways
Genes Cells, 18, 2013
|
|
3WA0
 
 | | Crystal structure of merlin complexed with DCAF1/VprBP | | Descriptor: | Merlin, Protein VPRBP | | Authors: | Mori, T, Gotoh, S, Shirakawa, M, Hakoshima, T. | | Deposit date: | 2013-04-20 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis of DDB1-and-Cullin 4-associated Factor 1 (DCAF1) recognition by merlin/NF2 and its implication in tumorigenesis by CD44-mediated inhibition of merlin suppression of DCAF1 function.
Genes Cells, 19, 2014
|
|
3X23
 
 | | Radixin complex | | Descriptor: | Peptide from Matrix metalloproteinase-14, Radixin | | Authors: | Terawaki, S, Kitano, K, Aoyama, M, Mori, T, Hakoshima, T. | | Deposit date: | 2014-12-09 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | MT1-MMP recognition by ERM proteins and its implication in CD44 shedding
Genes Cells, 20, 2015
|
|
