4NHJ
 
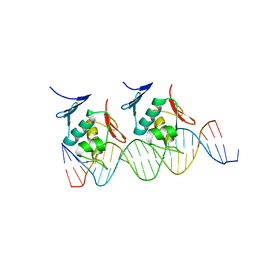 | | Crystal structure of Klebsiella pneumoniae RstA DNA-binding domain in complex with RstA box | | Descriptor: | 5'-D(*CP*AP*GP*GP*GP*AP*GP*TP*AP*AP*CP*GP*GP*AP*AP*TP*GP*TP*AP*CP*AP*AP*C)-3', 5'-D(*GP*GP*TP*TP*GP*TP*AP*CP*AP*TP*TP*CP*CP*GP*TP*TP*AP*CP*TP*CP*CP*CP*T)-3', DNA-binding transcriptional regulator RstA | | Authors: | Li, Y.C, Hsiao, C.D. | | Deposit date: | 2013-11-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural dynamics of the two-component response regulator RstA in recognition of promoter DNA element.
Nucleic Acids Res., 42, 2014
|
|
6LBI
 
 | |
5AWV
 
 | |
5EGM
 
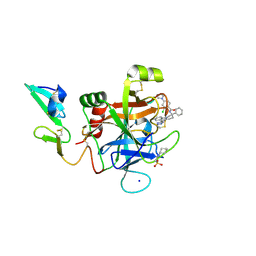 | | Development of a novel tricyclic class of potent and selective FIXa inhibitors | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-chloranyl-~{N}-[(7~{S})-2-methyl-7-phenyl-10-(1~{H}-1,2,3,4-tetrazol-5-yl)-8,9-dihydro-6~{H}-pyrido[1,2-a]indol-7-yl]-4-(1,2,4-triazol-4-yl)benzamide, Coagulation factor IX, ... | | Authors: | Meng, D, Andre, P, Bateman, T.J, Berger, R, Chen, Y, Desai, K, Dewnani, S, Ellsworth, K, Feng, D, Geissler, W.M, Guo, L, Hruza, A, Jian, T, Li, H, Parker, D.L, Reichert, P, Sherer, E.C, Smith, C.J, Sonatore, L.M, Tschirret-Guth, R, Wu, J, Xu, J, Zhang, T, Campeau, L, Orr, R, Poirier, M, McCabe-Dunn, j, Araki, K, Nishimura, T, Sakurada, I, Hirabayashi, T, Wood, H.B. | | Deposit date: | 2015-10-27 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Development of a novel tricyclic class of potent and selective FIXa inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5GPK
 
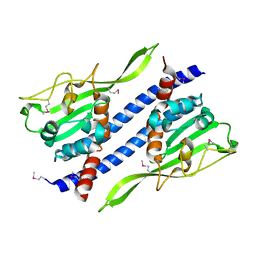 | | Crystal structure of Ccp1 mutant | | Descriptor: | Putative nucleosome assembly protein C36B7.08c | | Authors: | Yin, F, Gao, F, Chen, Y. | | Deposit date: | 2016-08-03 | | Release date: | 2016-11-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Ccp1 Homodimer Mediates Chromatin Integrity by Antagonizing CENP-A Loading
Mol.Cell, 64, 2016
|
|
7E9B
 
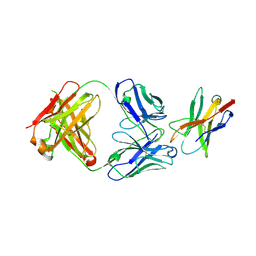 | |
3F4M
 
 | | Crystal structure of TIPE2 | | Descriptor: | CHLORIDE ION, Tumor necrosis factor, alpha-induced protein 8-like protein 2 | | Authors: | Zhang, X, Wang, J, Shi, Y. | | Deposit date: | 2008-11-01 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Crystal structure of TIPE2 provides insights into immune homeostasis
Nat.Struct.Mol.Biol., 16, 2009
|
|
3F8T
 
 | |
4X8C
 
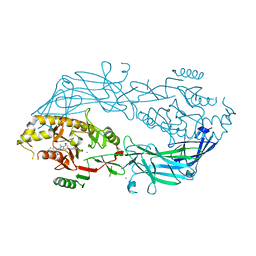 | | Crystal structure of human peptidylarginine deiminase type4 (PAD4) in complex with GSK147 | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type-4, [(3S,4R)-3-amino-4-hydroxypiperidin-1-yl]{2-[1-(cyclopropylmethyl)-1H-pyrrolo[2,3-b]pyridin-2-yl]-7-methoxy-1-methyl-1H-benzimidazol-5-yl}methanone | | Authors: | Lewis, H.D, Bax, B.D, Chung, C.-W, Polyakova, O, Thorpe, J. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation.
Nat.Chem.Biol., 11, 2015
|
|
4X8G
 
 | | Crystal structure of human peptidylarginine deiminase type4 (PAD4) in complex with GSK199 | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type-4, [(3R)-3-aminopiperidin-1-yl][2-(1-ethyl-1H-pyrrolo[2,3-b]pyridin-2-yl)-7-methoxy-1-methyl-1H-benzimidazol-5-yl]methanone | | Authors: | Lewis, H.D, Bax, B.D, Chung, C.-W, Polyakova, O, Thorpe, J. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation.
Nat.Chem.Biol., 11, 2015
|
|
9IVB
 
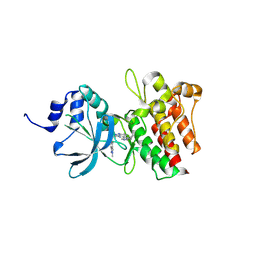 | |
4N6F
 
 | | Crystal structure of Amycolatopsis orientalis BexX complexed with G6P | | Descriptor: | CALCIUM ION, FRUCTOSE -6-PHOSPHATE, Putative thiosugar synthase | | Authors: | Zhang, X, Zhang, Y, Kinsland, C, Sasaki, E, Sun, H.G, Lu, M.J, Liu, T, Ou, A, Li, J, Chen, Y, Liu, H, Ealick, S.E. | | Deposit date: | 2013-10-11 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis.
Nature, 509, 2014
|
|
4N6E
 
 | | Crystal structure of Amycolatopsis orientalis BexX/CysO complex | | Descriptor: | Putative thiosugar synthase, SULFATE ION, ThiS/MoaD family protein | | Authors: | Zhang, X, Zhang, Y, Kinsland, C, Sasaki, E, Sun, H.G, Lu, M.J, Liu, T, Ou, A, Li, J, Chen, Y, Liu, H, Ealick, S.E. | | Deposit date: | 2013-10-11 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis.
Nature, 509, 2014
|
|
9CE4
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 indazole inhibitor compound 6 | | Descriptor: | (1S,4r)-4-[(1P)-5-chloro-1-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-6-yl]-1-(oxetan-3-yl)piperidin-1-ium, 1,2-ETHANEDIOL, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L, Zebisch, M, Henry, C, Barker, J.J. | | Deposit date: | 2024-06-26 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Discovery and Optimization of N-Heteroaryl Indazole LRRK2 Inhibitors.
J.Med.Chem., 67, 2024
|
|
4YZU
 
 | |
4Z0K
 
 | |
4ZAE
 
 | | Development of a novel class of potent and selective FIXa inhibitors | | Descriptor: | 2,6-dichloro-N-[(2R)-2-(5,6-dimethyl-1H-benzimidazol-2-yl)-2-phenylethyl]-4-(4H-1,2,4-triazol-4-yl)benzamide, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Coagulation factor IX, ... | | Authors: | Hruza, A, Reichert, P. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of a novel class of potent and selective FIXa inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8XAL
 
 | | Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein (Fragment), ... | | Authors: | Hsu, H.F, Wu, M.H, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2023-12-04 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Functional and structural investigation of a broadly neutralizing SARS-CoV-2 antibody.
JCI Insight, 9, 2024
|
|
8XBF
 
 | | Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, O5C2, heavy chain, ... | | Authors: | Hsu, H.F, Wu, M.H, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2023-12-06 | | Release date: | 2024-06-19 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional and structural investigation of a broadly neutralizing SARS-CoV-2 antibody.
JCI Insight, 9, 2024
|
|
4L72
 
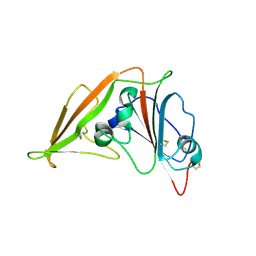 | | Crystal structure of MERS-CoV complexed with human DPP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Wang, X.Q, Wang, N.S. | | Deposit date: | 2013-06-13 | | Release date: | 2013-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4
Cell Res., 23, 2013
|
|
5Y3Q
 
 | |
5GPL
 
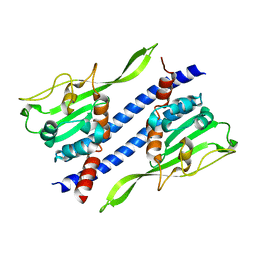 | | Crystal structure of Ccp1 | | Descriptor: | Putative nucleosome assembly protein C36B7.08c | | Authors: | Yin, F, Gao, F, Chen, Y. | | Deposit date: | 2016-08-03 | | Release date: | 2016-11-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ccp1 Homodimer Mediates Chromatin Integrity by Antagonizing CENP-A Loading
Mol.Cell, 64, 2016
|
|
7XUZ
 
 | | Crystal structure of a HDAC4-MEF2A-DNA ternary complex | | Descriptor: | DNA (5'-D(*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*A)-3'), Histone deacetylase 4, ... | | Authors: | Dai, S.Y, Guo, L, Dey, R, Guo, M, Bates, D, Cayford, J, Chen, X.J, Wei, X.D, Chen, L, Chen, Y.H. | | Deposit date: | 2022-05-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.591 Å) | | Cite: | Structural insights into the HDAC4-MEF2A-DNA complex and its implication in long-range transcriptional regulation.
Nucleic Acids Res., 52, 2024
|
|
7YBO
 
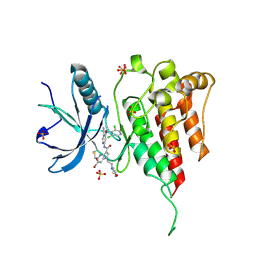 | | Crystal structure of FGFR4 kinase domain with 10z | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-6-bromanyl-5-(hydroxymethyl)-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YC3
 
 | | Crystal structure of FGFR4 kinase domain with 10t | | Descriptor: | 6-bromanyl-~{N}-[5-cyano-4-(2-methoxyethylamino)pyridin-2-yl]-5-methanoyl-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide, Fibroblast growth factor receptor 4, GLYCEROL, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.987 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
