5A3H
 
 | | 2-DEOXY-2-FLURO-B-D-CELLOBIOSYL/ENZYME INTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | ENDOGLUCANASE, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Davies, G.J, Varrot, A, Dauter, M, Brzozowski, A.M, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
1HFU
 
 | | TYPE-2 CU-DEPLETED LACCASE FROM COPRINUS CINEREUS at 1.68 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, LACCASE 1, ... | | Authors: | Ducros, V, Brzozowski, A.M. | | Deposit date: | 2000-12-08 | | Release date: | 2001-12-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of the Laccase from Coprinus Cinereus at 1.68A Resolution: Evidence for Different Type 2 Cu-Depleted Isoforms
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1A3H
 
 | |
3O0E
 
 | | Crystal structure of OmpF in complex with colicin peptide OBS1 | | Descriptor: | Colicin-E9, Porin OmpF, octyl beta-D-glucopyranoside | | Authors: | Wojdyla, J.A, Housden, N.G, Korczynska, J, Grishkovskaya, I, Kirkpatrick, N, Brzozowski, A.M, Kleanthous, C. | | Deposit date: | 2010-07-19 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Directed epitope delivery across the Escherichia coli outer membrane through the porin OmpF.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1QKN
 
 | |
1QKM
 
 | |
4A3H
 
 | | 2',4' DINITROPHENYL-2-DEOXY-2-FLURO-B-D-CELLOBIOSIDE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS AT 1.6 A RESOLUTION | | Descriptor: | 2,4-DINITROPHENYL-2-DEOXY-2-FLUORO-BETA-D-CELLOBIOSIDE, PROTEIN (ENDOGLUCANASE) | | Authors: | Davies, G.J, Brzozowski, A.M, Andersen, K, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-07-22 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
4OYL
 
 | |
4OYY
 
 | |
2FYI
 
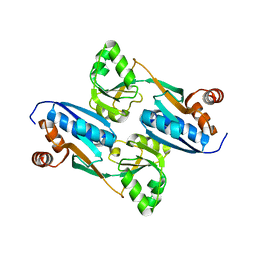 | | Crystal Structure of the Cofactor-Binding Domain of the Cbl Transcriptional Regulator | | Descriptor: | HTH-type transcriptional regulator cbl | | Authors: | Stec, E, Neumann, P, Wilkinson, A.J, Brzozowski, A.M, Bujacz, G.D. | | Deposit date: | 2006-02-08 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of the Sulphate Starvation Response in E. coli: Crystal Structure and Mutational Analysis of the Cofactor-binding Domain of the Cbl Transcriptional Regulator.
J.Mol.Biol., 364, 2006
|
|
1LV7
 
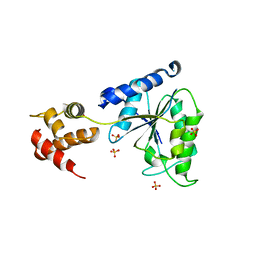 | | Crystal Structure of the AAA domain of FtsH | | Descriptor: | FtsH, SULFATE ION | | Authors: | Krzywda, S, Brzozowski, A.M, Verma, C, Karata, K, Ogura, T, Wilkinson, A.J. | | Deposit date: | 2002-05-26 | | Release date: | 2002-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the AAA domain of the ATP-dependent protease FtsH of Escherichia coli at 1.5 A resolution.
Structure, 10, 2002
|
|
1GWR
 
 | |
1HJ1
 
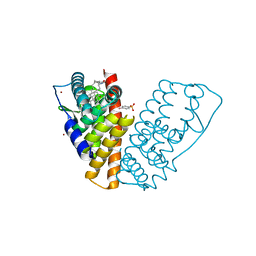 | | RAT OESTROGEN RECEPTOR BETA LIGAND-BINDING DOMAIN IN COMPLEX WITH PURE ANTIOESTROGEN ICI164,384 | | Descriptor: | N-BUTYL-11-[(7R,8R,9S,13S,14S,17S)-3,17-DIHYDROXY-13-METHYL-7,8,9,11,12,13,14,15,16,17-DECAHYDRO-6H-CYCLOPENTA[A]PHENANTHREN-7-YL]-N-METHYLUNDECANAMIDE, NICKEL (II) ION, OESTROGEN RECEPTOR BETA, ... | | Authors: | Pike, A.C.W, Brzozowski, A.M, Carlquist, M. | | Deposit date: | 2001-01-08 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights Into the Mode of Action of a Pure Antiestrogen
Structure, 9, 2001
|
|
1GWQ
 
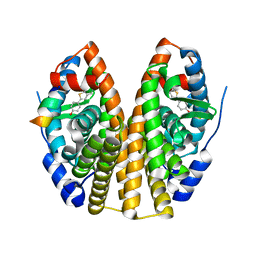 | |
1W8P
 
 | | Structural properties of the B25Tyr-NMe-B26Phe insulin mutant. | | Descriptor: | INSULIN A-CHAIN, INSULIN B-CHAIN, PHENOL, ... | | Authors: | Zakowa, L, Au-Alvarez, O, Dodson, E.J, Dodson, G.G, Brzozowski, A.M. | | Deposit date: | 2004-09-24 | | Release date: | 2005-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Towards the Insulin-Igf-I Intermediate Structures: Functional and Structural Properties of the B25Tyr-Nme-B26Phe Insulin Mutant.
Biochemistry, 43, 2004
|
|
1W9X
 
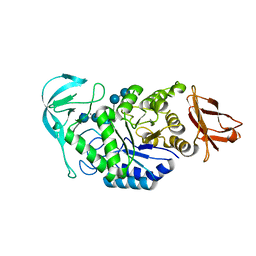 | | Bacillus halmapalus alpha amylase | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA AMYLASE, CALCIUM ION, ... | | Authors: | Davies, G.J, Brzozowski, A.M, Dauter, Z, Rasmussen, M.D, Borchert, T.V, Wilson, K.S. | | Deposit date: | 2004-10-20 | | Release date: | 2005-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Bacillus Halmapalus Family 13 Alpha-Amylase, Bha, in Complex with an Acarbose-Derived Nonasaccharide at 2.1 A Resolution
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1UTD
 
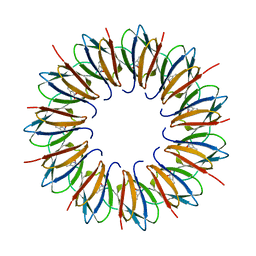 | | The structure of the trp RNA-binding attenuation protein (TRAP) bound to a 63-nucleotide RNA molecule containing GAGUUU repeats | | Descriptor: | 5'-R(*GP*UP*UP*UP*GP*AP)-3', TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Hopcroft, N.H, Manfredo, A, Wendt, A.L, Brzozowski, A.M, Gollnick, P, Antson, A.A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Interaction of RNA with Trap: The Role of Triplet Repeats and Separating Spacer Nucleotides
J.Mol.Biol., 338, 2004
|
|
1UOH
 
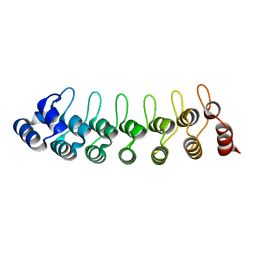 | | HUMAN GANKYRIN | | Descriptor: | 26S PROTEASOME NON-ATPASE REGULATORY SUBUNIT 10 | | Authors: | Krzywda, S, Brzozowski, A.M, Wilkinson, A.J. | | Deposit date: | 2003-09-17 | | Release date: | 2003-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Gankyrin, an Oncoprotein Found in Complexes with Cyclin-Dependent Kinase 4, a 19 S Proteasomal ATPase Regulator, and the Tumor Suppressors Rb and P53
J.Biol.Chem., 279, 2004
|
|
3W13
 
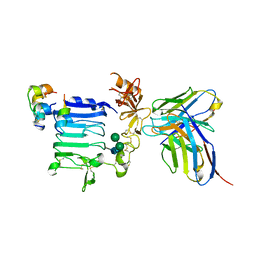 | | Insulin receptor ectodomain construct comprising domains L1-CR in complex with high-affinity insulin analogue [D-PRO-B26]-DTI-NH2, alphact peptide(693-719) and FAB 83-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, Insulin B chain, ... | | Authors: | Lawrence, M.C, Smith, B.J, Brzozowski, A.M. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.303 Å) | | Cite: | How insulin engages its primary binding site on the insulin receptor
Nature, 493, 2013
|
|
5FBZ
 
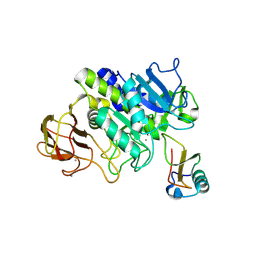 | | Structure of subtilase SubHal from Bacillus halmapalus - complex with chymotrypsin inhibitor CI2A | | Descriptor: | Autoproteolytic fragment of enzyme subtilase SubHal, CALCIUM ION, Enzyme subtilase SubHal from Bacillus halmapalus, ... | | Authors: | Dohnalek, J, Brzozowski, A.M, Svendsen, A, Wilson, K.S. | | Deposit date: | 2015-12-14 | | Release date: | 2016-05-18 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Stabilization of Enzymes by Metal Binding: Structures of Two Alkalophilic Bacillus Subtilases and Analysis of the Second Metal-Binding Site of the Subtilase Family
Book, 2016
|
|
5FAX
 
 | |
2A3H
 
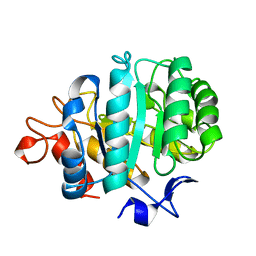 | | CELLOBIOSE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHERANS AT 2.0 A RESOLUTION | | Descriptor: | ENDOGLUCANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Davies, G.J, Brzozowski, A.M, Andersen, K, Schulein, M. | | Deposit date: | 1998-01-22 | | Release date: | 1999-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Bacillus agaradherans family 5 endoglucanase at 1.6 A and its cellobiose complex at 2.0 A resolution
Biochemistry, 37, 1998
|
|
2BSX
 
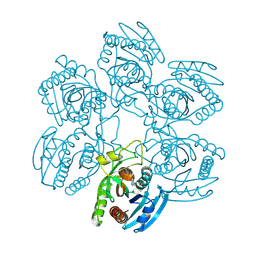 | | Crystal structure of the Plasmodium falciparum purine nucleoside phosphorylase complexed with inosine | | Descriptor: | INOSINE, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Schnick, C, Brzozowski, A.M, Dodson, E.J, Murshudov, G.N, Brannigan, J.A, Wilkinson, A.J. | | Deposit date: | 2005-05-24 | | Release date: | 2005-08-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Plasmodium Falciparum Purine Nucleoside Phosphorylase Complexed with Sulfate and its Natural Substrate Inosine
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2C0D
 
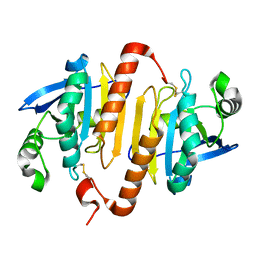 | | Structure of the mitochondrial 2-cys peroxiredoxin from Plasmodium falciparum | | Descriptor: | THIOREDOXIN PEROXIDASE 2 | | Authors: | Boucher, I.W, Brannigan, J.A, Wilkinson, A.J, Brzozowski, A.M, Muller, S. | | Deposit date: | 2005-09-01 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and Biochemical Characterisation of a Mitochondrial Peroxiredoxin from Plasmodium Falciparum
Mol.Microbiol., 61, 2006
|
|
2AAA
 
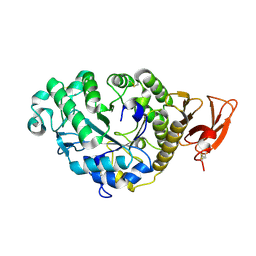 | | CALCIUM BINDING IN ALPHA-AMYLASES: AN X-RAY DIFFRACTION STUDY AT 2.1 ANGSTROMS RESOLUTION OF TWO ENZYMES FROM ASPERGILLUS | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION | | Authors: | Brady, L, Brzozowski, A.M, Derewenda, Z, Dodson, E.J, Dodson, G.G. | | Deposit date: | 1991-02-27 | | Release date: | 1993-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Calcium binding in alpha-amylases: an X-ray diffraction study at 2.1-A resolution of two enzymes from Aspergillus.
Biochemistry, 29, 1990
|
|
