7JXW
 
 | | EGFR kinase (T790M/V948R) in complex with osimertinib and JBJ-09-063 | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-{6-[4-(1-methylpiperidin-4-yl)phenyl]-1-oxo-1,3-dihydro-2H-isoindol-2-yl}-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors
Nat Commun, 13, 2022
|
|
7JXI
 
 | | EGFR kinase (T790M/V948R) in complex with PF-06747775 | | Descriptor: | Epidermal growth factor receptor, N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-methyl-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors
Nat Commun, 13, 2022
|
|
5OAC
 
 | |
5NXK
 
 | | L. reuteri 53608 SRRP | | Descriptor: | Serine-rich secreted cell wall anchored (LPXTG-motif ) protein | | Authors: | Sequeira, S, Dong, C. | | Deposit date: | 2017-05-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | Structural basis for the role of serine-rich repeat proteins from Lactobacillus reuteriin gut microbe-host interactions.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5NY0
 
 | | L. reuters 100-23 SRRP | | Descriptor: | L. reuteris SRRP binding region | | Authors: | Sequeira, S, Dong, C. | | Deposit date: | 2017-05-11 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the role of serine-rich repeat proteins from Lactobacillus reuteriin gut microbe-host interactions.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2JMX
 
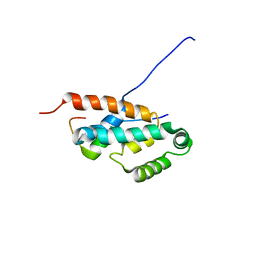 | | OSCP-NT (1-120) in complex with N-terminal (1-25) alpha subunit from F1-ATPase | | Descriptor: | ATP synthase O subunit, mitochondrial, ATP synthase subunit alpha heart isoform | | Authors: | Carbajo, R.J, Neuhaus, D, Kellas, F.A, Yang, J, Runswick, M.J, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2006-12-12 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | How the N-terminal Domain of the OSCP Subunit of Bovine F(1)F(o)-ATP Synthase Interacts with the N-terminal Region of an Alpha Subunit
J.Mol.Biol., 368, 2007
|
|
2JPS
 
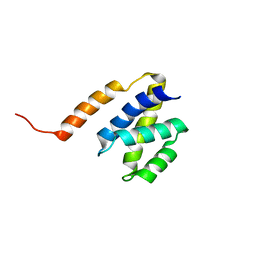 | | NAB2 N-terminal domain | | Descriptor: | Nuclear polyadenylated RNA-binding protein NAB2 | | Authors: | Grant, R, Marshall, N.J, Yang, J, Fasken, M, Kelly, S, Harreman, M.T, Neuhaus, D, Corbett, A.H, Stewart, M. | | Deposit date: | 2007-05-23 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal Mlp1-binding domain of the Saccharomyces cerevisiae mRNA-binding protein, Nab2
J.Mol.Biol., 376, 2008
|
|
1B4T
 
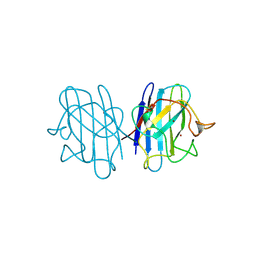 | | H48C YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-23 | | Release date: | 1999-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1B4L
 
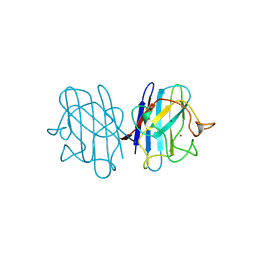 | | 15 ATMOSPHERE OXYGEN YEAST CU/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ZINC ION | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-22 | | Release date: | 1999-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1CPD
 
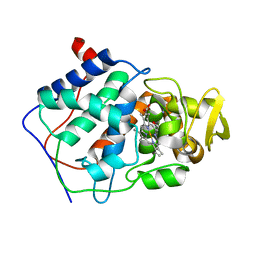 | |
1PY1
 
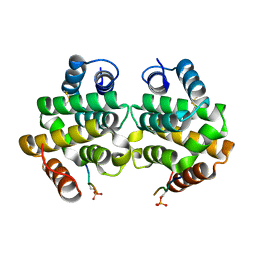 | | Complex of GGA1-VHS domain and beta-secretase C-terminal phosphopeptide | | Descriptor: | ADP-ribosylation factor binding protein GGA1, Beta-secretase | | Authors: | Zhu, G, Zhang, X.C. | | Deposit date: | 2003-07-07 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural characterization of the interaction of memapsin 2 (beta-secretase)
cytosolic domain with the VHS domain of GGA proteins.
Biochemistry, 42, 2003
|
|
4DXC
 
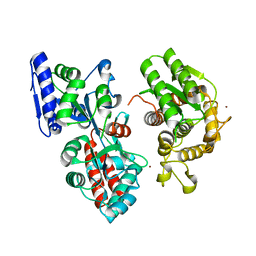 | |
5O2D
 
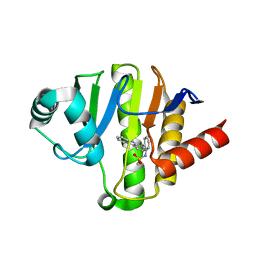 | | PARP14 Macrodomain 2 with inhibitor | | Descriptor: | Poly [ADP-ribose] polymerase 14, ~{N}-[2-(9~{H}-carbazol-1-yl)phenyl]methanesulfonamide | | Authors: | Uth, K, Schuller, M, Sieg, C, Wang, J, Krojer, T, Knapp, S, Riedels, K, Bracher, F, Edwards, A.M, Arrowsmith, C, Bountra, C, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-05-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a Selective Allosteric Inhibitor Targeting Macrodomain 2 of Polyadenosine-Diphosphate-Ribose Polymerase 14.
ACS Chem. Biol., 12, 2017
|
|
4JSK
 
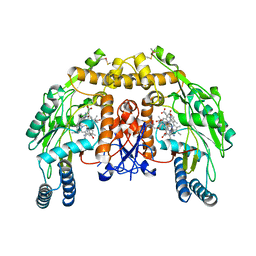 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 6,6'-(pentane-1,5-diyl)bis(4-methylpyridin-2-amine) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-pentane-1,5-diylbis(4-methylpyridin-2-amine), ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-03-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | In search of potent and selective inhibitors of neuronal nitric oxide synthase with more simple structures.
Bioorg.Med.Chem., 21, 2013
|
|
1CPF
 
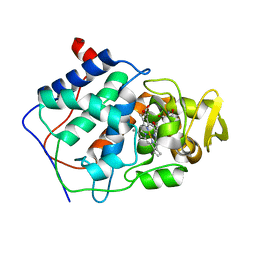 | | A CATION BINDING MOTIF STABILIZES THE COMPOUND I RADICAL OF CYTOCHROME C PEROXIDASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Miller, M.A, Han, G.W, Kraut, J. | | Deposit date: | 1994-08-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cation binding motif stabilizes the compound I radical of cytochrome c peroxidase.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1FAD
 
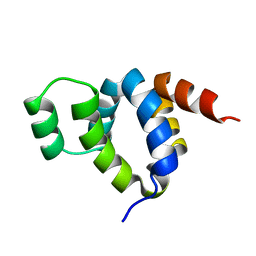 | | DEATH DOMAIN OF FAS-ASSOCIATED DEATH DOMAIN PROTEIN, RESIDUES 89-183 | | Descriptor: | PROTEIN (FADD PROTEIN) | | Authors: | Jeong, E.-J, Bang, S, Lee, T.H, Park, Y.-I, Sim, W.-S, Kim, K.-S. | | Deposit date: | 1999-03-23 | | Release date: | 1999-07-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of FADD death domain. Structural basis of death domain interactions of Fas and FADD.
J.Biol.Chem., 274, 1999
|
|
2KQC
 
 | | Second PBZ domain of human APLF protein | | Descriptor: | Aprataxin and PNK-like factor, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
2KQB
 
 | | First PBZ domain of human APLF protein | | Descriptor: | Aprataxin and PNK-like factor, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
1EER
 
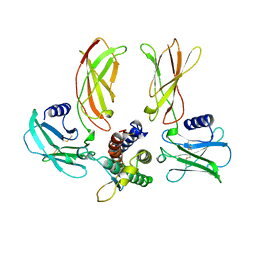 | |
5ORG
 
 | |
4DXB
 
 | |
1V11
 
 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, BENZAMIDINE, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain {Alpha}-Keto Acid Decarboxylase/Dehydrogenase
J.Biol.Chem., 279, 2004
|
|
2ER7
 
 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES.III. THREE-DIMENSIONAL STRUCTURE OF ENDOTHIAPEPSIN COMPLEXED WITH A TRANSITION-STATE ISOSTERE INHIBITOR OF RENIN AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | ENDOTHIAPEPSIN, SULFATE ION, TRANSITION-STATE ISOSTERE INHIBITOR OF RENIN | | Authors: | Veerapandian, B, Cooper, J.B, Szelke, M, Blundell, T.L. | | Deposit date: | 1990-11-12 | | Release date: | 1991-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray analyses of aspartic proteinases. III Three-dimensional structure of endothiapepsin complexed with a transition-state isostere inhibitor of renin at 1.6 A resolution.
J.Mol.Biol., 216, 1990
|
|
1V1M
 
 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, BENZAMIDINE, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-20 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain Alpha-Keto Acid Decarboxylase/Dehydrogenase.
J.Biol.Chem., 279, 2004
|
|
2KQD
 
 | | First PBZ domain of human APLF protein in complex with ribofuranosyladenosine | | Descriptor: | ADENOSINE, Aprataxin and PNK-like factor, ZINC ION, ... | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
