1AJA
 
 | |
5FKI
 
 | | Pseudorabies virus (PrV) nuclear egress complex proteins fitted as a hexameric lattice into a sub-tomogram average derived from focused- ion beam milled lamellae electron cryo-microscopic data | | Descriptor: | CHLORIDE ION, UL31, UL34 protein, ... | | Authors: | Hagen, C, Dent, K.C, Zeev Ben Mordehai, T, Vasishtan, D, Antonin, W, Mettenleiter, T.C, Gruenewald, K. | | Deposit date: | 2015-10-16 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights Into Inner Nuclear Membrane Remodelling
Cell Rep., 13, 2015
|
|
8QMP
 
 | | Structure of the E2 Beryllium Fluoride Complex of the Autoinhibited Calcium ATPase ACA8 | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Calcium-transporting ATPase 8, plasma membrane-type, ... | | Authors: | Thirup Larsen, S, Karlsen Dannersoe, J, Nissen, P. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conserved N-terminal Regulation of the ACA8 Calcium Pump with Two Calmodulin Binding Sites.
J.Mol.Biol., 436, 2024
|
|
3C6T
 
 | | Crystal Structure of HIV Reverse Transcriptase in complex with inhibitor 14 | | Descriptor: | 2-[3-chloro-5-(3-chloro-5-cyanophenoxy)phenoxy]-N-(2-chloro-4-sulfamoylphenyl)acetamide, Reverse transcriptase | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2008-02-05 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The design and synthesis of diaryl ether second generation HIV-1 non-nucleoside reverse transcriptase inhibitors (NNRTIs) with enhanced potency versus key clinical mutations.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1RAX
 
 | |
5E2T
 
 | | Crystal structure of anti-TAU antibody AT8 FAB | | Descriptor: | AT8 HEAVY CHAIN, AT8 LIGHT CHAIN, CALCIUM ION | | Authors: | Malia, T, Teplyakov, A. | | Deposit date: | 2015-10-01 | | Release date: | 2016-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Epitope mapping and structural basis for the recognition of phosphorylated tau by the anti-tau antibody AT8.
Proteins, 84, 2016
|
|
3C6U
 
 | | Crystal Structure of HIV Reverse Transcriptase in complex with inhibitor 22 | | Descriptor: | 3-chloro-5-[2-chloro-5-(1H-indazol-3-ylmethoxy)phenoxy]benzonitrile, Reverse transcriptase | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2008-02-05 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The design and synthesis of diaryl ether second generation HIV-1 non-nucleoside reverse transcriptase inhibitors (NNRTIs) with enhanced potency versus key clinical mutations.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5AQ5
 
 | |
8H2A
 
 | |
8H2B
 
 | | Crystal structure of alcohol dehydrogenase from Zobellia galactanivorans | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Brott, S, Bornscheuer, U.T, Nam, K.H. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unique alcohol dehydrogenases involved in algal sugar utilization by marine bacteria
Appl.Microbiol.Biotechnol., 107, 2023
|
|
3QUZ
 
 | | Structure of the mouse CD1d-NU-alpha-GalCer-iNKT TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Li, Y, Girardi, E, Yu, E.D, Zajonc, D.M. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis.
Embo J., 30, 2011
|
|
3QUY
 
 | | Structure of the mouse CD1d-BnNH-GSL-1'-iNKT TCR complex | | Descriptor: | (2S,3R,4S,5R,6S)-6-[(2S,3S,4R)-2-(hexacosanoylamino)-3,4-dihydroxy-octadecoxy]-3,4,5-trihydroxy-N-(phenylmethyl)oxane-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, Y, Girardi, E, Yu, E.D, Zajonc, D.M. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis.
Embo J., 30, 2011
|
|
4G56
 
 | | Crystal Structure of full length PRMT5/MEP50 complexes from Xenopus laevis | | Descriptor: | Hsl7 protein, MGC81050 protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Ho, M, Wilczek, C, Bonanno, J, Shechter, D, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity
Plos One, 8, 2013
|
|
5O5D
 
 | | Cellobiohydrolase Cel7A from T. atroviride | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Borisova, A.S, Stahlberg, J, Hansson, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Correlation of structure, function and protein dynamics in GH7 cellobiohydrolases from Trichoderma atroviride, T. reesei and T. harzianum.
Biotechnol Biofuels, 11, 2018
|
|
3QUX
 
 | | Structure of the mouse CD1d-alpha-C-GalCer-iNKT TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Li, Y, Girardi, E, Yu, E.D, Zajonc, D.M. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis.
Embo J., 30, 2011
|
|
5O59
 
 | | Cellobiohydrolase Cel7A from T. atroviride | | Descriptor: | 1-thio-beta-D-glucopyranose, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Borisova, A.S, Stahlberg, J, Hansson, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Correlation of structure, function and protein dynamics in GH7 cellobiohydrolases from Trichoderma atroviride, T. reesei and T. harzianum.
Biotechnol Biofuels, 11, 2018
|
|
2N8L
 
 | |
2N8M
 
 | |
2Z9Q
 
 | |
5U6D
 
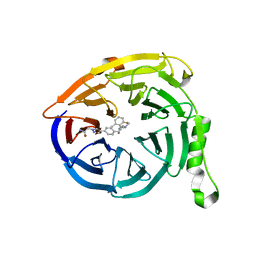 | |
5UXH
 
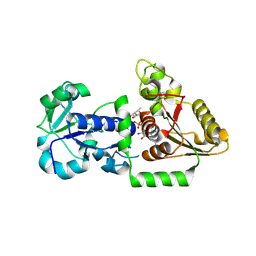 | | Structure of Human POFUT1 in complex with GDP-fucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE | | Authors: | Xu, X, McMillan, B, Blacklow, S.C. | | Deposit date: | 2017-02-22 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure of human POFUT1, its requirement in ligand-independent oncogenic Notch signaling, and functional effects of Dowling-Degos mutations.
Glycobiology, 27, 2017
|
|
2M4Q
 
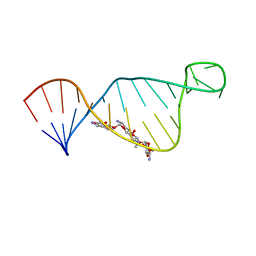 | | NMR structure of E. coli ribosomela decoding site with apramycin | | Descriptor: | APRAMYCIN, RNA (27-MER) | | Authors: | Puglisi, J.D, Tsai, A, Marshall, R, Viani, E. | | Deposit date: | 2013-02-10 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The impact of aminoglycosides on the dynamics of translation elongation.
Cell Rep, 3, 2013
|
|
5WNS
 
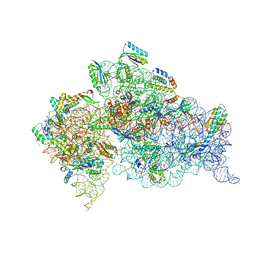 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S Ribosomal RNA rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | DeMirci, H. | | Deposit date: | 2017-08-01 | | Release date: | 2018-02-21 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 2'-O-methylation in mRNA disrupts tRNA decoding during translation elongation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5WNR
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S Ribosomal RNA rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | DeMirci, H. | | Deposit date: | 2017-08-01 | | Release date: | 2018-02-21 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 2'-O-methylation in mRNA disrupts tRNA decoding during translation elongation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2BQW
 
 | | CRYSTAL STRUCTURE OF FACTOR XA IN COMPLEX WITH COMPOUND 45 | | Descriptor: | 1-{2-[(4-CHLOROPHENYL)AMINO]-2-OXOETHYL}-N-(1-ISOPROPYLPIPERIDIN-4-YL)-1H-INDOLE-2-CARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR X, ... | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-28 | | Release date: | 2006-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
