4ES4
 
 | | Crystal structure of YdiV and FlhD complex | | Descriptor: | Flagellar transcriptional regulator FlhD, Putative cyclic di-GMP regulator CdgR | | Authors: | Li, B, Gu, L. | | Deposit date: | 2012-04-22 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility
Nucleic Acids Res., 40, 2012
|
|
8QX6
 
 | | Novel laminarin-binding CBM X584 | | Descriptor: | PKD domain-containing protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Zuehlke, M.K, Jeudy, A, Czjzek, M. | | Deposit date: | 2023-10-22 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unveiling the role of novel carbohydrate-binding modules in laminarin interaction of multimodular proteins from marine Bacteroidota during phytoplankton blooms.
Environ.Microbiol., 26, 2024
|
|
8SZY
 
 | |
7MHC
 
 | | Structure of human STING in complex with MK-1454 | | Descriptor: | (2R,5R,7R,8S,10R,12aR,14R,15S,15aR,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-15,16-difluoro-2,10-bis(sulfanyl)octahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A kinase-cGAS cascade to synthesize a therapeutic STING activator.
Nature, 603, 2022
|
|
4QXM
 
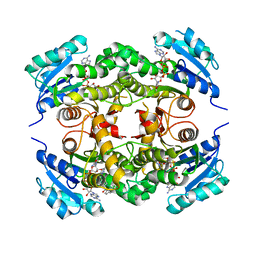 | | Crystal structure of the InhA:GSK_SB713 complex | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], N-(2-chloro-4-fluorobenzyl)-4-[(3,5-dimethyl-1H-pyrazol-1-yl)methyl]benzamide, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Gulten, G, Sacchettini, J.C. | | Deposit date: | 2014-07-21 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | N-Benzyl-4-((heteroaryl)methyl)benzamides: A New Class of Direct NADH-Dependent 2-trans Enoyl-Acyl Carrier Protein Reductase (InhA) Inhibitors with Antitubercular Activity.
Chemmedchem, 11, 2016
|
|
3DLZ
 
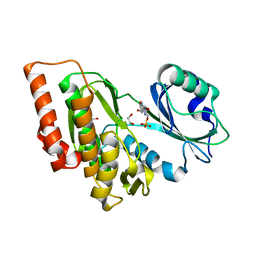 | | Crystal structure of human haspin in complex with AMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Keates, T, Burgess-Brown, N, Murray, J.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Wickstroem, M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-30 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and functional characterization of the atypical human kinase haspin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7DQ9
 
 | |
7C83
 
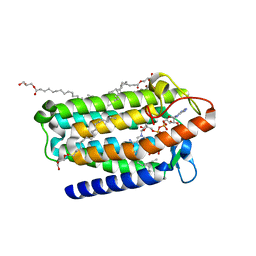 | | Crystal structure of an integral membrane steroid 5-alpha-reductase PbSRD5A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-oxo-5-alpha-steroid 4-dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ren, R.B, Han, Y.F, Xiao, Q.J, Deng, D. | | Deposit date: | 2020-05-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of steroid reductase SRD5A reveals conserved steroid reduction mechanism.
Nat Commun, 12, 2021
|
|
8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | Authors: | Zhao, R.C, Wu, L.L, Han, P. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3IQ7
 
 | | Crystal Structure of human Haspin in complex with 5-Iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, IODIDE ION, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Keates, T, Burgess-Brown, N, Fedorov, O, Pike, A.C.W, Von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-19 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and functional characterization of the atypical human kinase haspin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7KN3
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain complexed with a pre-pandemic antibody S-B8 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, S-B8 Fab heavy chain, ... | | Authors: | Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Neutralizing Antibodies to SARS-CoV-2 Selected from a Human Antibody Library Constructed Decades Ago.
Adv Sci, 9, 2022
|
|
7KN4
 
 | |
5YMV
 
 | | Crystal structure of 9-mer peptide from influenza virus in complex with BF2*1201 | | Descriptor: | ALA-VAL-LYS-GLY-VAL-GLY-THR-MET-VAL, Beta-2-microglobulin, Class I histocompatibility antigen, ... | | Authors: | Xiao, J, Xiang, W, Qi, J, Chai, Y, Liu, W.J, Gao, G.F. | | Deposit date: | 2017-10-22 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | An Invariant Arginine in Common with MHC Class II Allows Extension at the C-Terminal End of Peptides Bound to Chicken MHC Class I.
J Immunol., 201, 2018
|
|
7A9I
 
 | |
7A9J
 
 | |
5YMW
 
 | | Crystal structure of 8-mer peptide from Rous sarcoma virus in complex with BF2*1201 | | Descriptor: | Beta-2-microglobulin, Class I histocompatibility antigen, F10 alpha chain, ... | | Authors: | Xiao, J, Xiang, W, Qi, J, Chai, Y, Liu, W.J, Gao, G.F. | | Deposit date: | 2017-10-22 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | An Invariant Arginine in Common with MHC Class II Allows Extension at the C-Terminal End of Peptides Bound to Chicken MHC Class I.
J Immunol., 201, 2018
|
|
5YLS
 
 | | Crystal structure of T2R-TTL-Y50 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, E-3-(3-azanyl-4-methoxy-phenyl)-1-(5-methoxy-2,2-dimethyl-chromen-8-yl)prop-2-en-1-one, ... | | Authors: | Yang, J.H, Chen, L.J. | | Deposit date: | 2017-10-18 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 293, 2018
|
|
5H4B
 
 | | Crystal structure of Cbln4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-4 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5YLJ
 
 | | Crystal structure of T2R-TTL-Millepachine complex | | Descriptor: | (E)-1-(5-methoxy-2,2-dimethyl-chromen-8-yl)-3-(4-methoxyphenyl)prop-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Yang, J.H, Chen, L.J. | | Deposit date: | 2017-10-17 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 293, 2018
|
|
7DD2
 
 | |
7DK4
 
 | |
7DDN
 
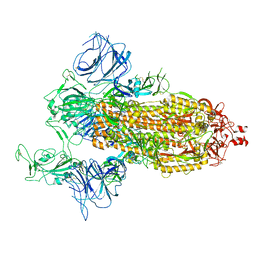 | | SARS-Cov2 S protein at open state | | Descriptor: | Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2020-10-29 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections.
Nat Commun, 12, 2021
|
|
7DDD
 
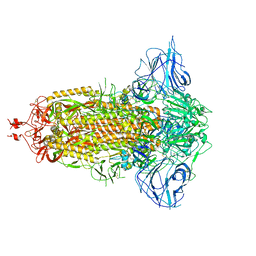 | | SARS-Cov2 S protein at close state | | Descriptor: | Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2020-10-28 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections.
Nat Commun, 12, 2021
|
|
7DCX
 
 | |
7DK7
 
 | |
