5W0L
 
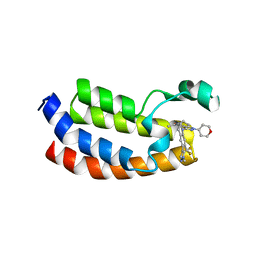 | | CREBBP Bromodomain in complex with Cpd10 (1-(3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydro-2H-pyran-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-(oxan-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-31 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
8JQK
 
 | | Crystal structure of a carbonyl reductase SSCR mutant from Sporobolomyces Salmonicolor | | Descriptor: | Aldehyde reductase 2 | | Authors: | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
8JQJ
 
 | | Crystal structure of carbonyl reductase SSCR mutant 1 from Sporobolomyces Salmonicolor | | Descriptor: | Aldehyde reductase 2 | | Authors: | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
7JQ3
 
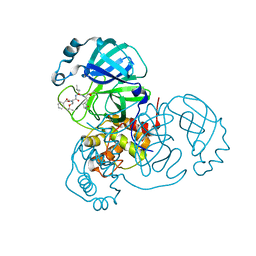 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI6 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JPZ
 
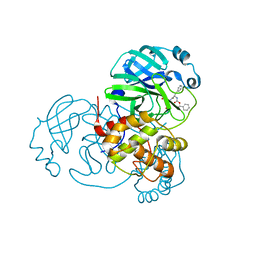 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI1 | | Descriptor: | (phenylmethyl) N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C-like proteinase | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ5
 
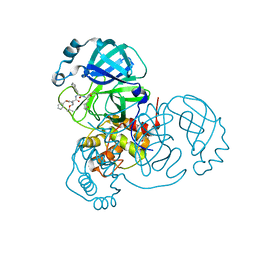 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI8 | | Descriptor: | 3C-like proteinase, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
5XWZ
 
 | | Crystal structure of a lactonase from Cladophialophora bantiana | | Descriptor: | GLYCEROL, MALONATE ION, SODIUM ION, ... | | Authors: | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-06-30 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization and crystal structure of a novel zearalenone hydrolase from Cladophialophora bantiana
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
7JQ1
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI4 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl]propan-2-yl}-L-phenylalaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
4MJ5
 
 | | Crystal Structure of HLA-A*1101 in complex with H1-22, an influenza A(H1N1) virus epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Liu, J, Tan, S, Zhao, M, Qi, J, Gao, G.F. | | Deposit date: | 2013-09-03 | | Release date: | 2014-10-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Cross-immunity Against Avian Influenza A(H7N9) Virus in the Healthy Population Is Affected by Antigenicity-Dependent Substitutions.
J.Infect.Dis., 214, 2016
|
|
6BB3
 
 | |
4MJ6
 
 | | Crystal Structure of HLA-A*1101 in complex with H7-22, an influenza A(H7N9) virus epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Liu, J, Tan, S, Zhao, M, Qi, J, Gao, G.F. | | Deposit date: | 2013-09-03 | | Release date: | 2014-10-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Cross-immunity Against Avian Influenza A(H7N9) Virus in the Healthy Population Is Affected by Antigenicity-Dependent Substitutions.
J.Infect.Dis., 214, 2016
|
|
6BB1
 
 | | Lactate Dehydrogenase in complex with inhibitor (R)-5-((2-chlorophenyl)thio)-6'-(4-fluorophenoxy)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro-[2,2'-bipyridin]-6(1H)-one | | Descriptor: | (2R)-5-[(2-chlorophenyl)sulfanyl]-6'-(4-fluorophenoxy)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro[2,2'-bipyridin]-6(1H)-one, L-lactate dehydrogenase A chain, LACTIC ACID, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided optimization and in vivo activities of hydroxylactone and hydroxylactam Inhibitors of Human Lactate Dehydrogenase
To Be Published
|
|
6BB2
 
 | | Lactate Dehydrogenase in complex with inhibitor (S)-5-((2-chlorophenyl)thio)-6'-(4-fluorophenoxy)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro-[2,2'-bipyridin]-6(1H)-one | | Descriptor: | (2S)-5-[(2-chlorophenyl)sulfanyl]-6'-(4-fluorophenoxy)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro[2,2'-bipyridin]-6(1H)-one, L-lactate dehydrogenase A chain, LACTIC ACID, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure-guided optimization and in vivo activities of hydroxylactone and hydroxylactam Inhibitors of Human Lactate Dehydrogenase
To Be Published
|
|
6BAG
 
 | | Lactate Dehydrogenase in complex with inhibitor (R)-5-((2-chlorophenyl)thio)-6'-((4-fluorophenyl)amino)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro-[2,2'-bipyridin]-6(1H)-one | | Descriptor: | (2R)-5-[(2-chlorophenyl)sulfanyl]-6'-[(4-fluorophenyl)amino]-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro[2,2'-bipyridin]-6(1H)-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, L-lactate dehydrogenase A chain, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-guided optimization and in vivo activities of hydroxylactone and hydroxylactam Inhibitors of Human Lactate Dehydrogenase
To Be Published
|
|
6BB0
 
 | |
6BAZ
 
 | | Lactate Dehydrogenase in complex with inhibitor (S)-5-((2-chlorophenyl)thio)-6'-((4-fluorophenyl)amino)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro-[2,2'-bipyridin]-6(1H)-one | | Descriptor: | (3S,6S)-3-[(2-chlorophenyl)sulfanyl]-6-{6-[(4-fluorophenyl)amino]pyridin-2-yl}-6-(thiophen-3-yl)piperidine-2,4-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, L-lactate dehydrogenase A chain, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-guided optimization and in vivo activities of hydroxylactone and hydroxylactam Inhibitors of Human Lactate Dehydrogenase
To Be Published
|
|
6BAD
 
 | |
6B21
 
 | | Crystal structure of AmtB from E. coli bound to TopFluor cardiolipin | | Descriptor: | Ammonia channel, [(2R,5R,11R,14S,24E)-14-[(acetyloxy)methyl]-8-{[5-(3,5-dimethyl-1H-pyrrol-2-yl-kappaN)-5-(3,5-dimethyl-2H-pyrrol-2-ylidene-kappaN)pentanoyl]oxy}-5,11-dihydroxy-2-{[(9E)-octadec-9-enoyl]oxy}-5,11,16-trioxo-4,6,10,12,15-pentaoxa-5lambda~5~,11lambda~5~-diphosphatritriacont-24-en-1-yl (9E)-octadec-9-enoatato](difluoro)boron | | Authors: | Boone, C.D, Laganowsky, A. | | Deposit date: | 2017-09-19 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Allostery revealed within lipid binding events to membrane proteins.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BAX
 
 | |
7K0V
 
 | | Crystal structure of bRaf in complex with inhibitor GNE-0749 | | Descriptor: | CHLORIDE ION, N-(3,3-dimethylbutyl)-N'-{2-fluoro-5-[(5-fluoro-3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]-4-methylphenyl}urea, Non-specific serine/threonine protein kinase | | Authors: | Yin, J, Eigenbrot, C.E, Wang, W. | | Deposit date: | 2020-09-06 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Targeting KRAS Mutant Cancers via Combination Treatment: Discovery of a 5-Fluoro-4-(3 H )-quinazolinone Aryl Urea pan-RAF Kinase Inhibitor.
J.Med.Chem., 64, 2021
|
|
8HE4
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, ~{N}-oxidanylnaphthalene-1-carboxamide | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HE2
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, tert-butyl N-[3-[[4-(oxidanylcarbamoyl)phenyl]methylamino]-3-oxidanylidene-propyl]carbamate | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HE1
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | BENZHYDROXAMIC ACID, Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HFA
 
 | |
8HF9
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-10 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
