4JL1
 
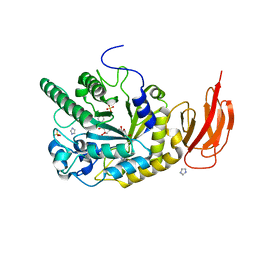 | | Crystal structure of a bacterial fucosidase with a multivalent iminocyclitol inhibitor | | Descriptor: | (3S,4R,5S)-N-benzyl-3,4-dihydroxy-5-methyl-D-prolinamide, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-03-12 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Exploring a Multivalent Approach to alpha-L-Fucosidase Inhibition
EUR.J.ORG.CHEM., 2013, 2013
|
|
6CXC
 
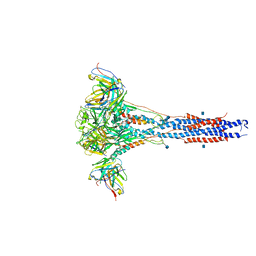 | | 3.9A Cryo-EM structure of murine antibody bound at a novel epitope of respiratory syncytial virus fusion protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, Envelope glycoprotein chimera, ... | | Authors: | Xie, Q, Wang, Z, Chen, X, Ni, F, Ma, J, Wang, Q. | | Deposit date: | 2018-04-02 | | Release date: | 2019-07-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure basis of neutralization by a novel site II/IV antibody against respiratory syncytial virus fusion protein.
Plos One, 14, 2019
|
|
2DTJ
 
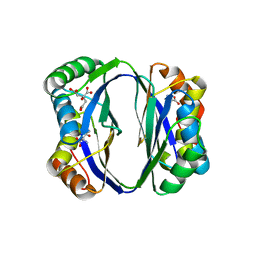 | | Crystal structure of regulatory subunit of aspartate kinase from Corynebacterium glutamicum | | Descriptor: | Aspartokinase, CITRIC ACID, THREONINE | | Authors: | Yoshida, A, Tomita, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2006-07-12 | | Release date: | 2007-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Insight into Concerted Inhibition of alpha(2)beta(2)-Type Aspartate Kinase from Corynebacterium glutamicum
J.Mol.Biol., 368, 2007
|
|
1XSO
 
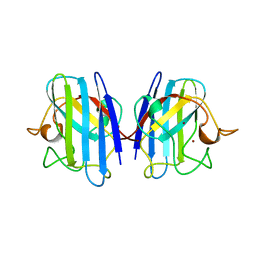 | | THREE-DIMENSIONAL STRUCTURE OF XENOPUS LAEVIS CU,ZN SUPEROXIDE DISMUTASE B DETERMINED BY X-RAY CRYSTALLOGRAPHY AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Djinovic Carugo, K, Coda, A, Battistoni, A, Carri, M.T, Polticelli, F, Desideri, A, Rotilio, G, Wilson, K.S, Bolognesi, M. | | Deposit date: | 1995-03-14 | | Release date: | 1995-07-10 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Three-dimensional structure of Xenopus laevis Cu,Zn superoxide dismutase b determined by X-ray crystallography at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
7BNU
 
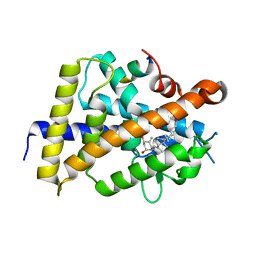 | | VDR complex with BXL-62 | | Descriptor: | 1,25-Dihydroxy-16-ene-20-cyclopropyl-vitamin D3, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Belorusova, A.Y. | | Deposit date: | 2021-01-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Vitamin D Analogs Bearing C-20 Modifications Stabilize the Agonistic Conformation of Non-Responsive Vitamin D Receptor Variants.
Int J Mol Sci, 23, 2022
|
|
7BNS
 
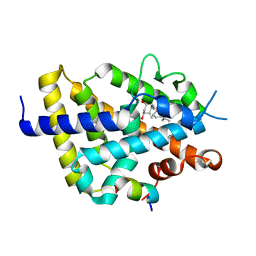 | | VDR complex with BXL-62 | | Descriptor: | 1,25-Dihydroxy-16-ene-20-cyclopropyl-vitamin D3, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Belorusova, A.Y. | | Deposit date: | 2021-01-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Vitamin D Analogs Bearing C-20 Modifications Stabilize the Agonistic Conformation of Non-Responsive Vitamin D Receptor Variants.
Int J Mol Sci, 23, 2022
|
|
8G4A
 
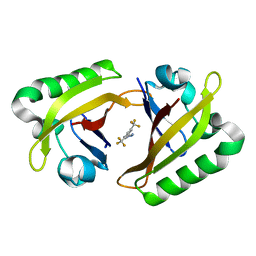 | |
1QLQ
 
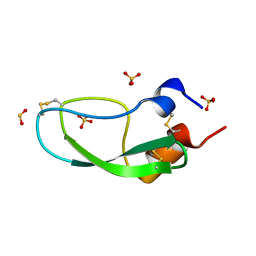 | | Bovine Pancreatic Trypsin Inhibitor (BPTI) Mutant with Altered Binding Loop Sequence | | Descriptor: | PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Czapinska, H, Krzywda, S, Sheldrick, G.M, Otlewski, J, Jaskolski, M. | | Deposit date: | 1999-09-10 | | Release date: | 1999-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High Resolution Structure of Bovine Pancreatic Trypsin Inhibitor with Altered Binding Loop Sequence
J.Mol.Biol., 295, 1999
|
|
3MKM
 
 | | Crystal structure of the E. coli pyrimidine nucleoside hydrolase YeiK (Apo-form) | | Descriptor: | CALCIUM ION, Putative uncharacterized protein YeiK | | Authors: | Garau, G, Fornili, A, Giabbai, B, Degano, M. | | Deposit date: | 2010-04-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Energy Landscapes Associated with Macromolecular Conformational Changes from Endpoint Structures
J.Am.Chem.Soc., 132, 2010
|
|
3D1H
 
 | |
4DHF
 
 | | Structure of Aurora A mutant bound to Biogenidec cpd 15 | | Descriptor: | 7-cyclopentyl-2-({1-methyl-5-[(4-methylpiperazin-1-yl)carbonyl]-1H-pyrrol-3-yl}amino)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A, MAGNESIUM ION, ... | | Authors: | Silvian, L, Marcotte, D.J. | | Deposit date: | 2012-01-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of 2,6,7-trisubstituted-7H-pyrrolo[2,3-d]pyrimidines as Aurora kinases inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3D1Q
 
 | | Structure of the PTP-Like Phytase Expressed by Selenomonas Ruminantium at an Ionic Strength of 400 mM | | Descriptor: | CHLORIDE ION, GLYCEROL, Myo-inositol hexaphosphate phosphohydrolase | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2008-05-06 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effect of ionic strength and oxidation on the P-loop conformation of the protein tyrosine phosphatase-like phytase, PhyAsr.
Febs J., 275, 2008
|
|
7KI6
 
 | |
7KI4
 
 | |
7BO6
 
 | | VDR complex with LCA derivative | | Descriptor: | (4R)-4-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2021-01-24 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Lithocholic acid-based design of noncalcemic vitamin D receptor agonists.
Bioorg.Chem., 111, 2021
|
|
2PT0
 
 | |
2PSZ
 
 | |
4KBS
 
 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 12:0 phosphatidic acid (12:0 PA) | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-ETHANEDIOL, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4QO2
 
 | | Crystal structure of rhomboid intramembrane protease GlpG in complex with peptide derived inhibitor Ac-IATA-cmk | | Descriptor: | 6-AMINO-2-METHYL-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, CHLORIDE ION, Rhomboid protease GlpG, ... | | Authors: | Zoll, S, Strisovsky, K. | | Deposit date: | 2014-06-19 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate binding and specificity of rhomboid intramembrane protease revealed by substrate-peptide complex structures.
Embo J., 33, 2014
|
|
5PTI
 
 | |
4MGS
 
 | | BiXyn10A CBM1 APO | | Descriptor: | Putative glycosyl hydrolase family 10 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2013-08-28 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2DWS
 
 | | Cu-containing nitrite reductase at pH 8.4 with bound nitrite | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Jacobson, F. | | Deposit date: | 2006-08-16 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | pH Dependence of Copper Geometry, Reduction Potential, and Nitrite Affinity in Nitrite Reductase
J.Biol.Chem., 282, 2007
|
|
4MGQ
 
 | | PbXyn10C CBM APO | | Descriptor: | CALCIUM ION, Glycosyl hydrolase family 10 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2013-08-28 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QO0
 
 | |
3RWQ
 
 | | Discovery of a Novel, Potent and Selective Inhibitor of 3-Phosphoinositide Dependent Kinase (PDK1) | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Kazmirski, S, Kohls, D. | | Deposit date: | 2011-05-09 | | Release date: | 2011-11-16 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of Novel, Potent, and Selective Inhibitors of 3-Phosphoinositide-Dependent Kinase (PDK1).
J.Med.Chem., 54, 2011
|
|
