8X7M
 
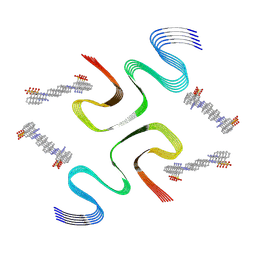 | | CR-bound E46K alpha-synuclein fibrils | | Descriptor: | 4-azanyl-3-[(~{E})-[4-[4-[(~{E})-(1-azanyl-4-sulfo-naphthalen-2-yl)diazenyl]phenyl]phenyl]diazenyl]naphthalene-1-sulfonic acid, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7L
 
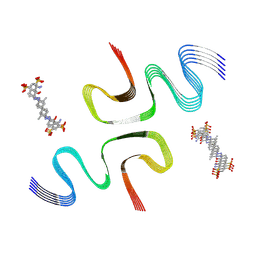 | | EB-bound E46K alpha-synuclein fibrils | | Descriptor: | 4-azanyl-6-[[4-[4-[(~{E})-(8-azanyl-1-oxidanyl-5,7-disulfo-naphthalen-2-yl)diazenyl]-3-methyl-phenyl]-2-methyl-phenyl]diazenyl]-5-oxidanyl-naphthalene-1,3-disulfonic acid, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6AW2
 
 | | Crystal structure of the HopQ-CEACAM1 complex | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, HopQ | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
6AVZ
 
 | | Crystal structure of the HopQ-CEACAM3 WT complex | | Descriptor: | CALCIUM ION, Carcinoembryonic antigen-related cell adhesion molecule 3, HopQ, ... | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
5F22
 
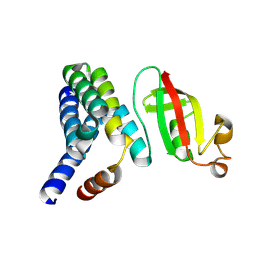 | | C-terminal domain of SARS-CoV nsp8 complex with nsp7 | | Descriptor: | Non-structural protein, Non-structural protein 7 | | Authors: | Li, S. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-27 | | Last modified: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | C-terminal domain of SARS-CoV nsp8 complex with nsp7
To Be Published
|
|
6AW1
 
 | | Crystal structure of CEACAM3 | | Descriptor: | CHLORIDE ION, Carcinoembryonic antigen-related cell adhesion molecule 3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
6KOX
 
 | |
8K4H
 
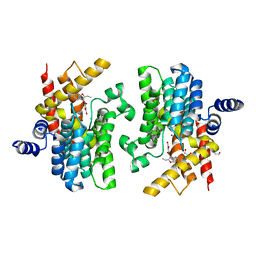 | | Crystal structure of PDE4D complexed with benzbromarone | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, ZINC ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-07-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8K4C
 
 | | Crystal structure of PDE4D complexed with ethaverine hydrochloride | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3,4-diethoxyphenyl)methyl]-6,7-diethoxy-isoquinoline, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8H77
 
 | | Hsp90-AhR-p23-XAP2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, Aryl hydrocarbon receptor, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
5HZW
 
 | | Crystal structure of the orphan region of human endoglin/CD105 in complex with BMP9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Growth/differentiation factor 2, Maltose-binding periplasmic protein,Endoglin, ... | | Authors: | Bokhove, M, Saito, T, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.451 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
7W1S
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-007 | | Descriptor: | Nanobody Nb-007, Spike protein S1 | | Authors: | Yang, J, Lin, S, Sun, H.L, Lu, G.W. | | Deposit date: | 2021-11-20 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | A Potent Neutralizing Nanobody Targeting the Spike Receptor-Binding Domain of SARS-CoV-2 and the Structural Basis of Its Intimate Binding.
Front Immunol, 13, 2022
|
|
5I05
 
 | |
7D32
 
 | | The TBA-Pb2+ complex in P41212 space group | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
7D31
 
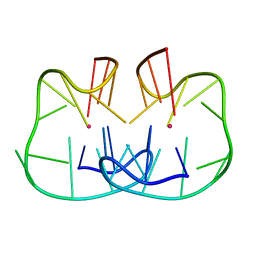 | | The TBA-Pb2+ complex in P41212 space group | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
7D33
 
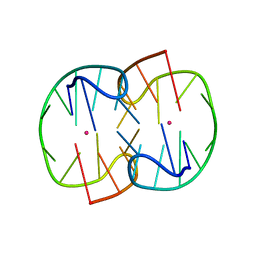 | | The Pb2+ complexed structure of TBA G8C mutant | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*CP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
7W1Y
 
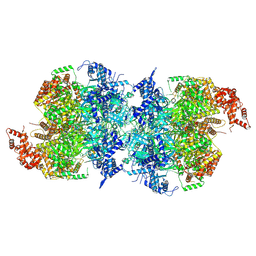 | | Human MCM double hexamer bound to natural DNA duplex (polyAT/polyTA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (49-MER), ... | | Authors: | Li, J, Dong, J, Dang, S, Zhai, Y. | | Deposit date: | 2021-11-21 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | The human pre-replication complex is an open complex.
Cell, 186, 2023
|
|
7W26
 
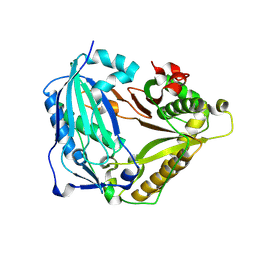 | | monolignol ferulate transferase | | Descriptor: | Ferulate monolignol transferase | | Authors: | Xi, L, Shuliu, D, Yue, F, Yi, Z. | | Deposit date: | 2021-11-22 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of the plant feruloyl-coenzyme A monolignol transferase provides insights into the formation of monolignol ferulate conjugates.
Biochem.Biophys.Res.Commun., 594, 2022
|
|
7C43
 
 | |
7C4B
 
 | | The crystal structure of Trypanosoma brucei RNase D : UMP complex | | Descriptor: | CCHC-type domain-containing protein, MANGANESE (II) ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Gao, Y.Q, Gan, J.H. | | Deposit date: | 2020-05-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural basis for guide RNA trimming by RNase D ribonuclease in Trypanosoma brucei.
Nucleic Acids Res., 49, 2021
|
|
7C4C
 
 | | The crystal structure of Trypanosoma brucei RNase D : GMP complex | | Descriptor: | CCHC-type domain-containing protein, GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Gao, Y.Q, Gan, J.H. | | Deposit date: | 2020-05-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | Structural basis for guide RNA trimming by RNase D ribonuclease in Trypanosoma brucei.
Nucleic Acids Res., 49, 2021
|
|
7C45
 
 | |
7C47
 
 | | The crystal structure of Trypanosoma brucei RNase D : CMP complex | | Descriptor: | CCHC-type domain-containing protein, CYTIDINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Gao, Y.Q, Gan, J.H. | | Deposit date: | 2020-05-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for guide RNA trimming by RNase D ribonuclease in Trypanosoma brucei.
Nucleic Acids Res., 49, 2021
|
|
7C42
 
 | | The crystal structure of Trypanosoma brucei RNase D | | Descriptor: | CCHC-type domain-containing protein, ZINC ION | | Authors: | Gao, Y.Q, Gan, J.H. | | Deposit date: | 2020-05-14 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for guide RNA trimming by RNase D ribonuclease in Trypanosoma brucei.
Nucleic Acids Res., 49, 2021
|
|
8IGZ
 
 | | Xcc NAMPT Quadruple mutant | | Descriptor: | Pre-B cell enhancing factor related protein | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2023-02-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural insights into Xanthomonas campestris pv. campestris NAD + biosynthesis via the NAM salvage pathway.
Commun Biol, 7, 2024
|
|
