4LWB
 
 | | Structure of Bacillus subtilis nitric oxide synthase in complex with 6-((((3R,5S)-5-(((6-amino-4-methylpyridin-2-yl)methoxy)methyl)pyrrolidin-3-yl)oxy)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-({[(3R,5S)-5-{[(6-amino-4-methylpyridin-2-yl)methoxy]methyl}pyrrolidin-3-yl]oxy}methyl)-4-methylpyridin-2-amine, CHLORIDE ION, ... | | Authors: | Holden, J.K, Li, H, Poulos, T.L. | | Deposit date: | 2013-07-26 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and biological studies on bacterial nitric oxide synthase inhibitors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LUW
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 6-((((3R,5S)-5-(((6-amino-4-methylpyridin-2-yl)methoxy)methyl)pyrrolidin-3-yl)oxy)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-({[(3R,5S)-5-{[(6-amino-4-methylpyridin-2-yl)methoxy]methyl}pyrrolidin-3-yl]oxy}methyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-07-25 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and biological studies on bacterial nitric oxide synthase inhibitors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LWA
 
 | | Structure of Bacillus subtilis nitric oxide synthase in complex with ((2S, 3S)-1,3-bis((6-(2,5-dimethyl-1H-pyrrol-1-yl)-4-methylpyridin-2-yl)methoxy)-2-aminobutane | | Descriptor: | 6,6'-{[(2S,3S)-2-aminobutane-1,3-diyl]bis(oxymethanediyl)}bis(4-methylpyridin-2-amine), CHLORIDE ION, GLYCEROL, ... | | Authors: | Holden, J.K, Li, H, Poulos, T.P. | | Deposit date: | 2013-07-26 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and biological studies on bacterial nitric oxide synthase inhibitors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3DN5
 
 | | Aldose Reductase in complex with novel biarylic inhibitor | | Descriptor: | 3-[5-(3-nitrophenyl)thiophen-2-yl]propanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Klebe, G. | | Deposit date: | 2008-07-01 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Based Optimization of Aldose Reductase Inhibitors Originating from Virtual Screening
Chemmedchem, 4, 2009
|
|
1R5A
 
 | | Glutathione S-transferase | | Descriptor: | COPPER (II) ION, GLUTATHIONE SULFONIC ACID, glutathione transferase | | Authors: | Oakley, A.J. | | Deposit date: | 2003-10-09 | | Release date: | 2003-10-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification, characterization and structure of a new Delta class glutathione transferase isoenzyme.
Biochem.J., 388, 2005
|
|
4LUX
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-((((3R,5S)-5-(((6-amino-4-methylpyridin-2-yl)methoxy)methyl)pyrrolidin-3-yl)oxy)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-({[(3R,5S)-5-{[(6-amino-4-methylpyridin-2-yl)methoxy]methyl}pyrrolidin-3-yl]oxy}methyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-07-25 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and biological studies on bacterial nitric oxide synthase inhibitors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1V2A
 
 | | Glutathione S-transferase 1-6 from Anopheles dirus species B | | Descriptor: | GLUTATHIONE SULFONIC ACID, glutathione transferase gst1-6 | | Authors: | Oakley, A.J. | | Deposit date: | 2003-10-10 | | Release date: | 2003-10-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification, characterization and structure of a new Delta class glutathione transferase isoenzyme.
Biochem.J., 388, 2005
|
|
1O3S
 
 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Chen, S, Ebright, R.H, Berman, H.M. | | Deposit date: | 2003-03-18 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex: Alteration of DNA Binding Specificity Through Alteration of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
7K7A
 
 | | Transmembrane structure of TNFR1 | | Descriptor: | Tumor necrosis factor receptor superfamily member 1A | | Authors: | Zhao, L, Chou, J. | | Deposit date: | 2020-09-22 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Diversity and Similarity of Transmembrane Trimerization of TNF Receptors.
Front Cell Dev Biol, 8, 2020
|
|
3RIZ
 
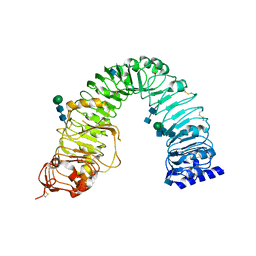 | | Crystal structure of the plant steroid receptor BRI1 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein BRASSINOSTEROID INSENSITIVE 1, ... | | Authors: | Hothorn, M. | | Deposit date: | 2011-04-14 | | Release date: | 2011-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.523 Å) | | Cite: | Structural basis of steroid hormone perception by the receptor kinase BRI1.
Nature, 474, 2011
|
|
3RJ0
 
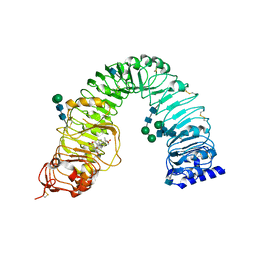 | | Plant steroid receptor BRI1 ectodomain in complex with brassinolide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinolide, Protein BRASSINOSTEROID INSENSITIVE 1, ... | | Authors: | Hothorn, M. | | Deposit date: | 2011-04-14 | | Release date: | 2011-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Structural basis of steroid hormone perception by the receptor kinase BRI1.
Nature, 474, 2011
|
|
3T4Q
 
 | |
3T4K
 
 | |
3T4O
 
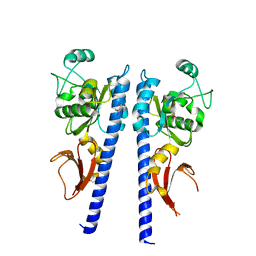 | |
3T4T
 
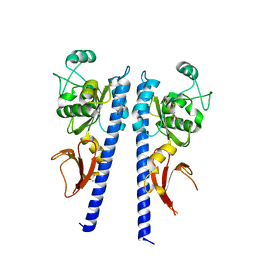 | |
3T4S
 
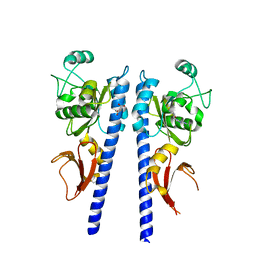 | |
3T4L
 
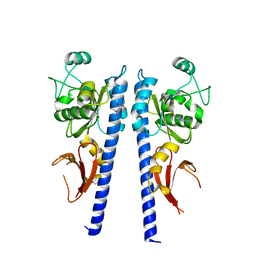 | |
3T4J
 
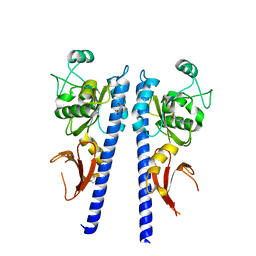 | |
1QUZ
 
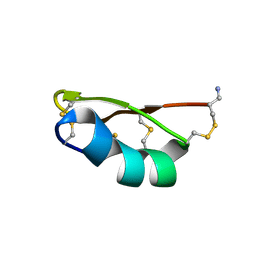 | | Solution structure of the potassium channel scorpion toxin HSTX1 | | Descriptor: | HSTX1 TOXIN | | Authors: | Savarin, P, Romi-Lebrun, R, Zinn-Justin, S, Lebrun, B, Nakajima, T, Gilquin, B, Menez, A. | | Deposit date: | 1999-07-05 | | Release date: | 2000-07-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural and functional consequences of the presence of a fourth disulfide bridge in the scorpion short toxins: solution structure of the potassium channel inhibitor HsTX1.
Protein Sci., 8, 1999
|
|
2R6Q
 
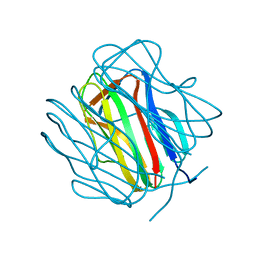 | |
2WYA
 
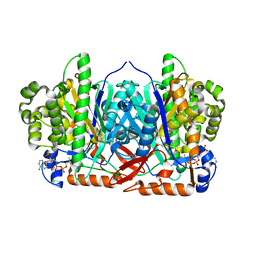 | | CRYSTAL STRUCTURE OF HUMAN MITOCHONDRIAL 3-HYDROXY-3-METHYLGLUTARYL- COENZYME A SYNTHASE 2 (HMGCS2) | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, GLYCEROL, HYDROXYMETHYLGLUTARYL-COA SYNTHASE, ... | | Authors: | Yue, W.W, Shafqat, N, Savitsky, P, Roos, A.K, Cooper, C, Murray, J.W, von Delft, F, Arrowsmith, C, Wikstrom, M, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2009-11-13 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Human Hmg-Coa Synthase Isoforms Provide Insights Into Inherited Ketogenesis Disorders and Inhibitor Design.
J.Mol.Biol., 398, 2010
|
|
