7DUF
 
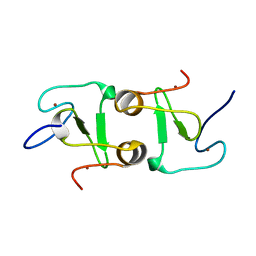 | | Crystal structure of VIM1 PHD finger. | | Descriptor: | E3 ubiquitin-protein ligase ORTHRUS 2, ZINC ION | | Authors: | Abhishek, S, Deeksha, W, Patel, D.J, Rajakumara, E. | | Deposit date: | 2021-01-08 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Helical and beta-Turn Conformations in the Peptide Recognition Regions of the VIM1 PHD Finger Abrogate H3K4 Peptide Recognition.
Biochemistry, 60, 2021
|
|
6J23
 
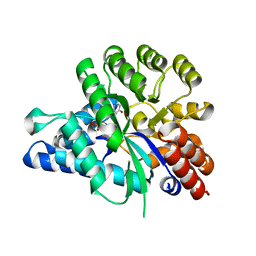 | | Crystal structure of arabidopsis ADAL complexed with GMP | | Descriptor: | Adenosine/AMP deaminase family protein, GUANOSINE-5'-MONOPHOSPHATE, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2018-12-30 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
6J4T
 
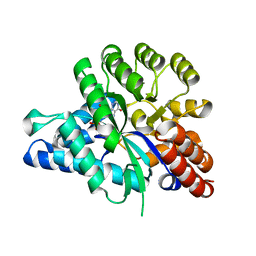 | | Crystal structure of arabidopsis ADAL complexed with IMP | | Descriptor: | Adenosine/AMP deaminase family protein, INOSINIC ACID, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
7LMM
 
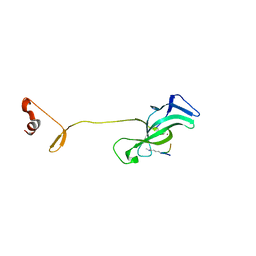 | |
7LMK
 
 | |
7W27
 
 | | Crystal structure of BEND3-BEN4-DNA complex | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(P*GP*GP*AP*CP*CP*CP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*GP*CP*TP*GP*CP*GP*TP*GP*GP*GP*TP*C)-3') | | Authors: | Zheng, L, Ren, A. | | Deposit date: | 2021-11-22 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Distinct structural bases for sequence-specific DNA binding by mammalian BEN domain proteins.
Genes Dev., 36, 2022
|
|
6AF0
 
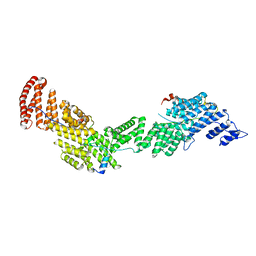 | | Structure of Ctr9, Paf1 and Cdc73 ternary complex from Myceliophthora thermophila | | Descriptor: | Cdc73 protein, Ctr9 protein, Paf1 protein | | Authors: | Wang, Z, Deng, P, Zhou, Y. | | Deposit date: | 2018-08-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Transcriptional elongation factor Paf1 core complex adopts a spirally wrapped solenoidal topology.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5HH7
 
 | |
2GDI
 
 | |
3NCU
 
 | |
4QBS
 
 | |
4TN7
 
 | | Crystal structure of mouse KDM2A-H3K36ME-NO complex | | Descriptor: | FE (III) ION, Lysine-specific demethylase 2A, NITRIC OXIDE, ... | | Authors: | Cheng, Z. | | Deposit date: | 2014-06-03 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A molecular threading mechanism underlies Jumonji lysine demethylase KDM2A regulation of methylated H3K36.
Genes Dev., 28, 2014
|
|
4QBR
 
 | |
3DIM
 
 | |
3DJ2
 
 | |
3F2W
 
 | |
3F4H
 
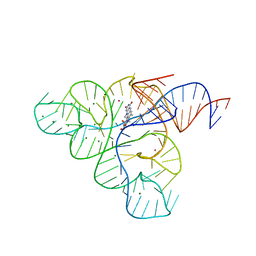 | | Crystal structure of the FMN riboswitch bound to roseoflavin | | Descriptor: | 1-deoxy-1-[8-(dimethylamino)-7-methyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl]-D-ribitol, FMN riboswitch, MAGNESIUM ION, ... | | Authors: | Serganov, A.A, Huang, L. | | Deposit date: | 2008-10-31 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch.
Nature, 458, 2009
|
|
3F2Q
 
 | |
3F4G
 
 | |
3F2X
 
 | |
3F2Y
 
 | |
3F30
 
 | |
3F2T
 
 | |
3DIO
 
 | |
3DIQ
 
 | |
