1NSX
 
 | |
7ERQ
 
 | |
7ERP
 
 | |
7FDF
 
 | |
6KBV
 
 | |
6KBO
 
 | | Three-dimensional LPS bound structure of VG16KRKP-KYE28. | | Descriptor: | Heparin cofactor 2, VG16KRKP | | Authors: | Ilyas, H, Bhunia, A. | | Deposit date: | 2019-06-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the combinatorial effects of antimicrobial peptides reveal a role of aromatic-aromatic interactions in antibacterial synergism.
J.Biol.Chem., 294, 2019
|
|
1NSR
 
 | |
1NSM
 
 | |
1NSZ
 
 | |
1NS2
 
 | |
1NSV
 
 | |
1NSS
 
 | |
1T36
 
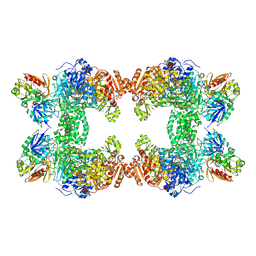 | | Crystal structure of E. coli carbamoyl phosphate synthetase small subunit mutant C248D complexed with uridine 5'-monophosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Carbamoyl-phosphate synthase large chain, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 2004-04-24 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Long-range allosteric transitions in carbamoyl phosphate synthetase.
Protein Sci., 13, 2004
|
|
4CRO
 
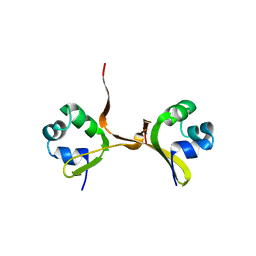 | | PROTEIN-DNA CONFORMATIONAL CHANGES IN THE CRYSTAL STRUCTURE OF A LAMBDA CRO-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*TP*CP*AP*CP*CP*GP*CP*GP*GP*GP*TP*GP*AP*TP*A)-3'), PROTEIN (LAMBDA CRO) | | Authors: | Brennan, R.G, Roderick, S.L, Takeda, Y, Matthews, B.W. | | Deposit date: | 1992-01-15 | | Release date: | 1992-01-15 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Protein-DNA conformational changes in the crystal structure of a lambda Cro-operator complex.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
1HXP
 
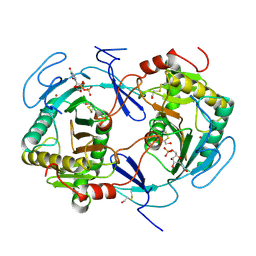 | | NUCLEOTIDE TRANSFERASE | | Descriptor: | BETA-MERCAPTOETHANOL, FE (III) ION, HEXOSE-1-PHOSPHATE URIDYLYLTRANSFERASE, ... | | Authors: | Wedekind, J.E, Frey, P.A, Rayment, I. | | Deposit date: | 1995-06-09 | | Release date: | 1996-11-08 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of galactose-1-phosphate uridylyltransferase from Escherichia coli at 1.8 A resolution.
Biochemistry, 34, 1995
|
|
1HXQ
 
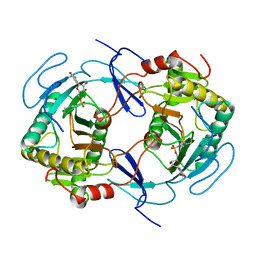 | | THE STRUCTURE OF NUCLEOTIDYLATED GALACTOSE-1-PHOSPHATE URIDYLYLTRANSFERASE FROM ESCHERICHIA COLI AT 1.86 ANGSTROMS RESOLUTION | | Descriptor: | FE (III) ION, HEXOSE-1-PHOSPHATE URIDYLYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Wedekind, J.E, Frey, P.A, Rayment, I. | | Deposit date: | 1996-06-16 | | Release date: | 1997-10-22 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of nucleotidylated histidine-166 of galactose-1-phosphate uridylyltransferase provides insight into phosphoryl group transfer.
Biochemistry, 35, 1996
|
|
1K6Q
 
 | | Crystal structure of antibody Fab fragment D3 | | Descriptor: | immunoglobulin Fab D3, heavy chain, light chain | | Authors: | Faelber, K, Kelley, R.F, Kirchhofer, D, Muller, Y.A. | | Deposit date: | 2001-10-17 | | Release date: | 2002-04-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of murine Fab D3 at 2.4 A resolution in comparison with the humanised Fab D3h44 (1.85A) provides structural insight into the humanisation process of the D3 anti-tissue factor antibody
To be Published
|
|
