8JH5
 
 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the Naproxen-bound state | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, Auxin efflux carrier component 1, nanobody | | Authors: | Sun, L, Liu, X, Yang, Z, Xia, J. | | Deposit date: | 2023-05-22 | | Release date: | 2024-05-29 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Chemical inhibition of Arabidopsis PIN-FORMED auxin transporters by the anti-inflammatory drug naproxen.
Plant Commun., 4, 2023
|
|
6IGA
 
 | |
8ILR
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 16 | | Descriptor: | N-[(2S)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8K0Z
 
 | | ABCG25 Wild Type purified with DDM in the apo-state | | Descriptor: | ABC transporter G family member 25 | | Authors: | Sun, L, Liu, X, Ying, W, Liao, L, Wei, H. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for abscisic acid efflux mediated by ABCG25 in Arabidopsis thaliana.
Nat.Plants, 9, 2023
|
|
8K0X
 
 | | ABCG25 Wild Type purified with DDM in the ABA-bound state | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABC transporter G family member 25 | | Authors: | Sun, L, Liu, X, Ying, W, Liao, L, Wei, H. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for abscisic acid efflux mediated by ABCG25 in Arabidopsis thaliana.
Nat.Plants, 9, 2023
|
|
6KGH
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis (apo-form) | | Descriptor: | COBALT (II) ION, Penicillin-binding protein PbpB, SODIUM ION | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-11 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGT
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with faropenem | | Descriptor: | (2R)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-5-[(2R)-oxolan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.308 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGU
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGV
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with amoxicillin | | Descriptor: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGS
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGW
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6L02
 
 | |
7WNQ
 
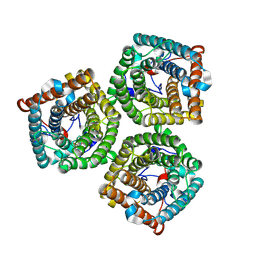 | | Cryo-EM structure of AtSLAC1 S59A mutant | | Descriptor: | Guard cell S-type anion channel SLAC1 | | Authors: | Sun, L, Liu, X, Li, Y. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the Arabidopsis guard cell anion channel SLAC1 suggests activation mechanism by phosphorylation.
Nat Commun, 13, 2022
|
|
7BZH
 
 | |
8ZPX
 
 | |
8ZQ4
 
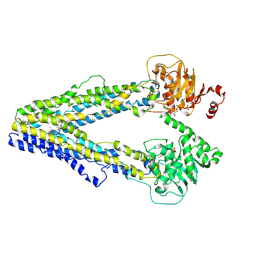 | | Structure of the Arabidopsis ABCB1 EQ mutant in the BL plus ATP bound state | | Descriptor: | ABC transporter B family member 1, ADENOSINE-5'-TRIPHOSPHATE, Brassinolide | | Authors: | Ying, W, Wei, H, Liu, X, Sun, L. | | Deposit date: | 2024-05-31 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into brassinosteroid export mediated by the Arabidopsis ABC transporter ABCB1.
Plant Commun., 6, 2025
|
|
8ZPZ
 
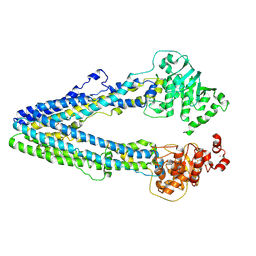 | | Structure of the wild-type Arabidopsis ABCB1 in the brassinolide-bound state | | Descriptor: | ABC transporter B family member 1, Brassinolide | | Authors: | Ying, W, Wei, H, Liu, X, Sun, L. | | Deposit date: | 2024-05-31 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into brassinosteroid export mediated by the Arabidopsis ABC transporter ABCB1.
Plant Commun., 6, 2025
|
|
7WC4
 
 | | Crystal structure of serotonin 2A receptor in complex with serotonin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7VP8
 
 | | Crystal structure of ferritin from Ureaplasma urealyticum | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin-like diiron domain-containing protein | | Authors: | Wang, W, Liu, X, Wang, Y, Fu, D, Wang, H. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Distinct structural characteristics define a new subfamily of Mycoplasma ferritin
Chin.Chem.Lett., 33, 2022
|
|
7WC5
 
 | | Crystal structure of serotonin 2A receptor in complex with psilocin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-(dimethylamino)ethyl]-1~{H}-indol-4-ol, 5-hydroxytryptamine receptor 2A, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC6
 
 | | Crystal structure of serotonin 2A receptor in complex with LSD | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC9
 
 | | Crystal structure of serotonin 2A receptor in complex with non-hallucinogenic psychedelic analog | | Descriptor: | (10~{R},15~{S})-12-[3-(2-methoxyphenyl)propyl]-4-methyl-1,4,12-triazatetracyclo[7.6.1.0^{5,16}.0^{10,15}]hexadeca-5(16),6,8-triene, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC7
 
 | | Crystal structure of serotonin 2A receptor in complex with lisuride | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC8
 
 | | Crystal structure of serotonin 2A receptor in complex with lumateperone | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(4-fluorophenyl)-4-[(10~{R},15~{S})-4-methyl-1,4,12-triazatetracyclo[7.6.1.0^{5,16}.0^{10,15}]hexadeca-5,7,9(16)-trien-12-yl]butan-1-one, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WZB
 
 | | lipopolysaccharide assembly protein LapB (open) | | Descriptor: | Lipopolysaccharide assembly protein B, TRIETHYLENE GLYCOL, ZINC ION | | Authors: | Yan, L, Dong, H, Li, H, Liu, X, Deng, Z, Dong, C, Zhang, Z. | | Deposit date: | 2022-02-17 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | lipopolysaccharide assembly protein LapB (open)
To Be Published
|
|
