2AOT
 
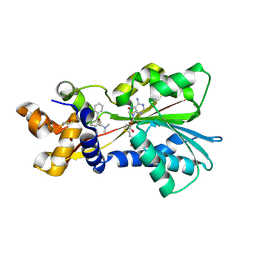 | | Histamine Methyltransferase Complexed with the Antihistamine Drug Diphenhydramine | | Descriptor: | Histamine N-methyltransferase, N-[2-(BENZHYDRYLOXY)ETHYL]-N,N-DIMETHYLAMINE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Cheng, X. | | Deposit date: | 2005-08-14 | | Release date: | 2005-09-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition of histamine N-methyltransferase by diverse drugs
J.Mol.Biol., 353, 2005
|
|
1G55
 
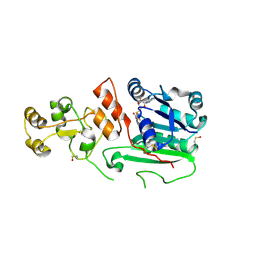 | | Structure of human DNMT2, an enigmatic DNA methyltransferase homologue | | Descriptor: | BETA-MERCAPTOETHANOL, DNA CYTOSINE METHYLTRANSFERASE DNMT2, GLYCEROL, ... | | Authors: | Dong, A, Yoder, J.A, Zhang, X, Zhou, L, Bestor, T.H, Cheng, X. | | Deposit date: | 2000-10-30 | | Release date: | 2001-01-17 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA.
Nucleic Acids Res., 29, 2001
|
|
1F3L
 
 | |
5KL7
 
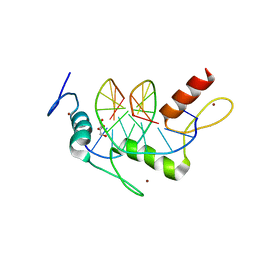 | | Wilms Tumor Protein (WT1) ZnF2-4Q369R in complex with carboxylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(1CC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
1YF3
 
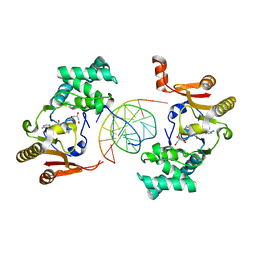 | | T4Dam in Complex with AdoHcy and 13-mer Oligonucleotide Making Non- and Semi-specific (~1/4) Contact | | Descriptor: | 5'-D(*AP*CP*CP*AP*TP*GP*AP*TP*CP*TP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*AP*GP*AP*TP*CP*AP*TP*GP*G)-3', DNA adenine methylase, ... | | Authors: | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | Deposit date: | 2004-12-30 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YFL
 
 | | T4Dam in Complex with Sinefungin and 16-mer Oligonucleotide Showing Semi-specific and Specific Contact and Flipped Base | | Descriptor: | 5'-D(P*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*GP*A)-3', DNA adenine methylase, SINEFUNGIN | | Authors: | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | Deposit date: | 2005-01-03 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
1CSN
 
 | | BINARY COMPLEX OF CASEIN KINASE-1 WITH MGATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CASEIN KINASE-1, MAGNESIUM ION, ... | | Authors: | Xu, R.-M, Cheng, X. | | Deposit date: | 1995-04-25 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of casein kinase-1, a phosphate-directed protein kinase.
EMBO J., 14, 1995
|
|
1YFJ
 
 | | T4Dam in Complex with AdoHcy and 15-mer Oligonucleotide Showing Semi-specific and Specific Contact | | Descriptor: | 5'-D(*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*G)-3', CALCIUM ION, CHLORIDE ION, ... | | Authors: | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | Deposit date: | 2005-01-02 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
1ORH
 
 | | Structure of the Predominant Protein Arginine Methyltransferase PRMT1 | | Descriptor: | GLYCEROL, Protein arginine N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhang, X, Cheng, X. | | Deposit date: | 2003-03-13 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of the Predominant Protein Arginine Methyltransferase PRMT1 and Analysis of Its Binding to Substrate Peptides
Structure, 11, 2003
|
|
1PEG
 
 | | Structural basis for the product specificity of histone lysine methyltransferases | | Descriptor: | Histone H3, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Zhang, X, Yang, Z, Khan, S.I, Horton, J.R, Tamaru, H, Selker, E.U, Cheng, X. | | Deposit date: | 2003-05-21 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for the product specificity of histone lysine methyltransferases
Mol.Cell, 12, 2003
|
|
1JQE
 
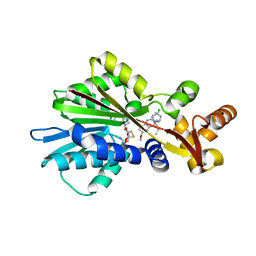 | | Crystal Structure Analysis of Human Histamine Methyltransferase (Ile105 Polymorphic Variant) Complexed with AdoHcy and Antimalarial Drug Quinacrine | | Descriptor: | Histamine N-Methyltransferase, QUINACRINE, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Zhang, X, Cheng, X. | | Deposit date: | 2001-08-06 | | Release date: | 2002-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Two polymorphic forms of human histamine methyltransferase: structural, thermal, and kinetic comparisons.
Structure, 9, 2001
|
|
1NGN
 
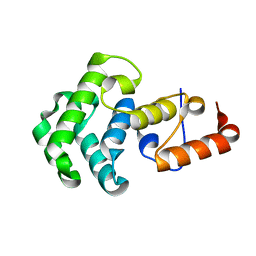 | | Mismatch repair in methylated DNA. Structure of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4 | | Descriptor: | methyl-CpG binding protein MBD4 | | Authors: | Wu, P, Qiu, C, Sohail, A, Zhang, X, Bhagwat, A.S, Cheng, X. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mismatch repair in methylated DNA. Structure and activity of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4
J.Biol.Chem., 278, 2003
|
|
5ISL
 
 | |
5IVV
 
 | |
1JQD
 
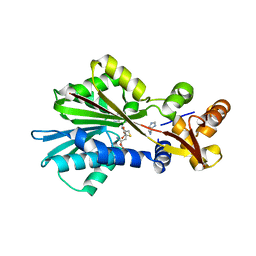 | | Crystal Structure Analysis of Human Histamine Methyltransferase (Thr105 Polymorphic Variant) Complexed with AdoHcy and Histamine | | Descriptor: | HISTAMINE, Histamine N-Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Zhang, X, Cheng, X. | | Deposit date: | 2001-08-06 | | Release date: | 2002-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Two polymorphic forms of human histamine methyltransferase: structural, thermal, and kinetic comparisons.
Structure, 9, 2001
|
|
1Q0S
 
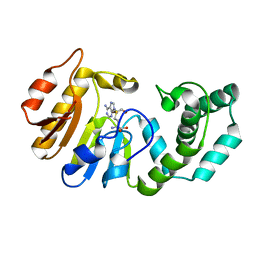 | | Binary Structure of T4DAM with AdoHcy | | Descriptor: | DNA adenine methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yang, Z, Horton, J.R, Zhou, L, Zhang, X.J, Dong, A, Zhang, X, Schlagman, S.L, Kossykh, V, Hattman, S, Cheng, X. | | Deposit date: | 2003-07-17 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the bacteriophage T4 DNA adenine methyltransferase
Nat.Struct.Biol., 10, 2003
|
|
5IVE
 
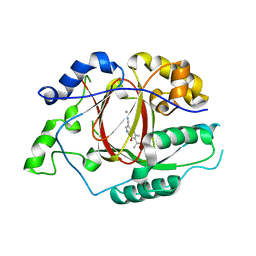 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N8 ( 5-methyl-7-oxo-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile) | | Descriptor: | 5-methyl-7-oxo-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, Lysine-specific demethylase 5A, MANGANESE (II) ION | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
1KHC
 
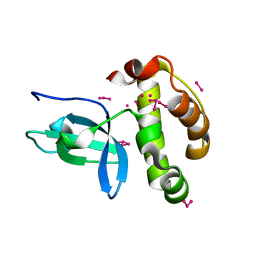 | | Crystal Structure of the PWWP Domain of Mammalian DNA Methyltransferase Dnmt3b | | Descriptor: | DNA cytosine-5 methyltransferase 3B2, UNKNOWN ATOM OR ION | | Authors: | Qiu, C, Sawada, K, Zhang, X, Cheng, X. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds.
Nat.Struct.Biol., 9, 2002
|
|
5IW0
 
 | |
1ORI
 
 | | Structure of the predominant protein arginine methyltransferase PRMT1 | | Descriptor: | Protein arginine N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN LIGAND | | Authors: | Zhang, X, Cheng, X. | | Deposit date: | 2003-03-13 | | Release date: | 2003-05-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Predominant Protein Arginine Methyltransferase PRMT1 and Analysis of its Binding to Substrate Peptides
Structure, 11, 2003
|
|
5IVC
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N3 (4'-[(2-phenylethyl)carbamoyl][2,2'-bipyridine]-4-carboxylic acid) | | Descriptor: | 1,2-ETHANEDIOL, 4'-[(2-phenylethyl)carbamoyl][2,2'-bipyridine]-4-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IWF
 
 | |
5IVB
 
 | |
5IVF
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N10 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol | | Descriptor: | 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol, Lysine-specific demethylase 5A, MANGANESE (II) ION | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IVJ
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N11 [3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid] | | Descriptor: | 3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-21 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.569 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
