3ROC
 
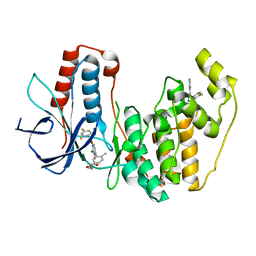 | | Crystal structure of human p38 alpha complexed with a pyrimidinone compound | | Descriptor: | 3-{5-chloro-4-[(2,4-difluorobenzyl)oxy]-6-oxopyrimidin-1(6H)-yl}-N-(2-hydroxyethyl)-4-methylbenzamide, 4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Shieh, H.-S, Xing, L. | | Deposit date: | 2011-04-25 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substituted N-aryl-6-pyrimidinones: A new class of potent, selective, and orally active p38 MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
8JXT
 
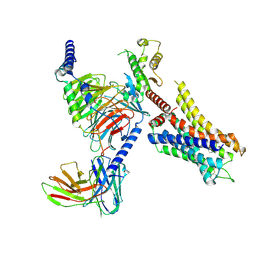 | | Histamine-bound H4R/Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2023-07-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of ligand recognition and design of antihistamines targeting histamine H 4 receptor.
Nat Commun, 15, 2024
|
|
8JXX
 
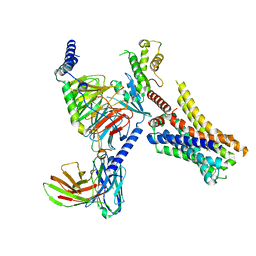 | | Clobenpropit-bound H4R/Gi complex | | Descriptor: | 3-(1~{H}-imidazol-4-yl)propyl ~{N}'-[(4-chlorophenyl)methyl]carbamimidothioate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2023-07-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of ligand recognition and design of antihistamines targeting histamine H 4 receptor.
Nat Commun, 15, 2024
|
|
8JXW
 
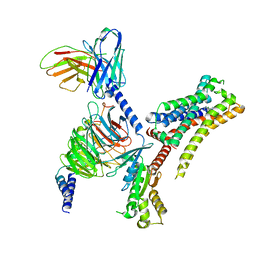 | | VUF6884-bound H4R/Gi complex | | Descriptor: | 2-chloranyl-6-(4-methylpiperazin-1-yl)benzo[b][1,4]benzoxazepine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2023-07-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis of ligand recognition and design of antihistamines targeting histamine H 4 receptor.
Nat Commun, 15, 2024
|
|
8JXV
 
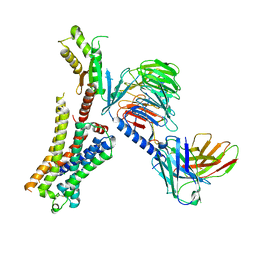 | | Clozapine-bound H4R/Gi complex | | Descriptor: | 3-chloranyl-6-(4-methylpiperazin-1-yl)-11~{H}-benzo[b][1,4]benzodiazepine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2023-07-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of ligand recognition and design of antihistamines targeting histamine H 4 receptor.
Nat Commun, 15, 2024
|
|
8HJ2
 
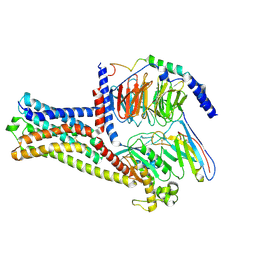 | | GPR21 wt with G15 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Chen, B, Lin, X, Xu, F. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
8HJ0
 
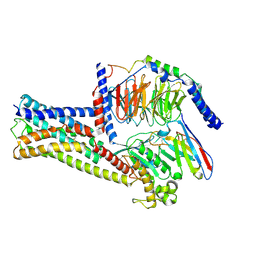 | | GPR21(m5) and G15 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Chen, B, Lin, X. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
8HIX
 
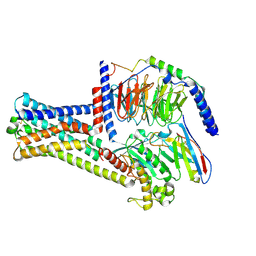 | | Cryo-EM structure of GPR21_m5_Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Chen, B, Lin, X, Xu, F. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
4LWT
 
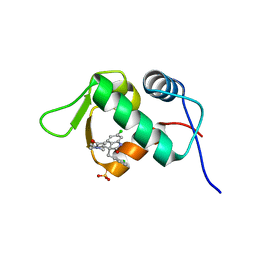 | | The 1.6A Crystal Structure of Humanized Xenopus MDM2 with RO5027344 | | Descriptor: | (3S)-3-[(3R)-1-acetylpiperidin-3-yl]-6-chloro-3-(3-chlorobenzyl)-1,3-dihydro-2H-indol-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C, Kammlott, U. | | Deposit date: | 2013-07-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of potent and selective spiroindolinone MDM2 inhibitor, RO8994, for cancer therapy.
Bioorg.Med.Chem., 22, 2014
|
|
3NZU
 
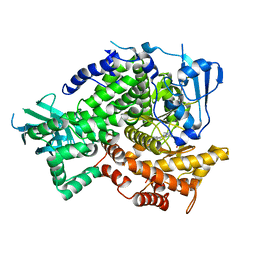 | |
3NZS
 
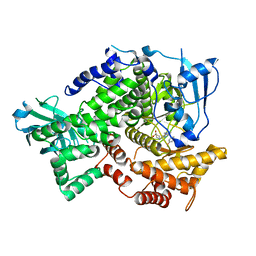 | |
6OOR
 
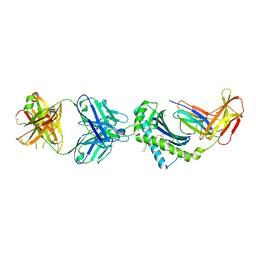 | | Structure of 1B1 bound to mouse CD1d | | Descriptor: | Antibody 1B1 Heavy chain, Antibody 1B1 Light chain, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Ying, G, Zajonc, D.M. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of NKT cell inhibition using the T-cell receptor-blocking anti-CD1d antibody 1B1.
J.Biol.Chem., 294, 2019
|
|
5Y18
 
 | |
3O50
 
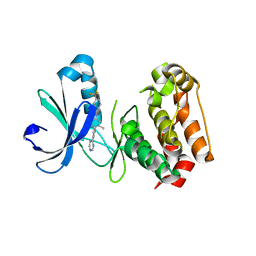 | | Crystal structure of benzamide 9 bound to AuroraA | | Descriptor: | N-{3-methyl-4-[(3-pyrimidin-4-ylpyridin-2-yl)oxy]phenyl}-3-(trifluoromethyl)benzamide, cDNA FLJ58295, highly similar to Serine/threonine-protein kinase 6 | | Authors: | Huang, X. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a potent, selective, and orally bioavailable pyridinyl-pyrimidine phthalazine aurora kinase inhibitor.
J.Med.Chem., 53, 2010
|
|
3VWB
 
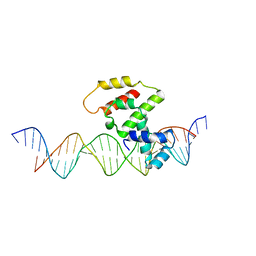 | | Crystal structure of VirB core domain (Se-Met derivative) complexed with the cis-acting site (5-BRU modifications) upstream icsb promoter | | Descriptor: | DNA (26-MER), Virulence regulon transcriptional activator virB | | Authors: | Gao, X.P, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2012-08-21 | | Release date: | 2013-08-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes
Nucleic Acids Res., 41, 2013
|
|
5YAG
 
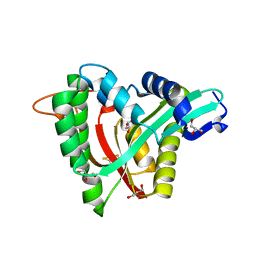 | | Crystal structure of mosquito arylalkylamine N-Acetyltransferase like 5b/spermine N-Acetyltransferase | | Descriptor: | AAEL004827-PA, GLYCEROL | | Authors: | Han, Q, Guan, H, Robinson, H, Li, J. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of aaNAT5b as a spermine N-acetyltransferase in the mosquito, Aedes aegypti.
PLoS ONE, 13, 2018
|
|
4LCZ
 
 | | Crystal structure of a multilayer-packed major light-harvesting complex | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wan, T, Li, M, Chang, W.R. | | Deposit date: | 2013-06-24 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a multilayer packed major light-harvesting complex: implications for grana stacking in higher plants.
Mol Plant, 7, 2014
|
|
1F03
 
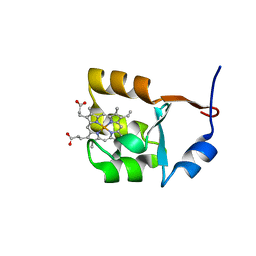 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
1F04
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
6OQO
 
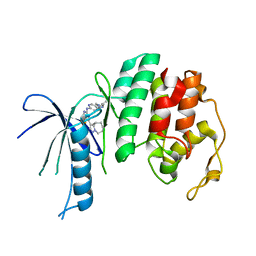 | | CDK6 in complex with Cpd24 N-(5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl)-5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)pyrimidin-2-amine | | Descriptor: | Cyclin-dependent kinase 6, N-[5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl]-5-fluoro-4-[(4R)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine | | Authors: | Murray, J.M, Boenig, G.D.L. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6OQL
 
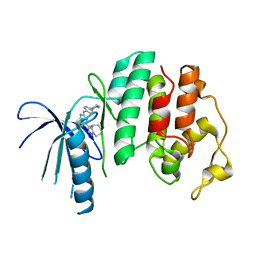 | | CDK6 in complex with Cpd13 (R)-5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)-N-(5-(4-methylpiperazin-1-yl)pyridin-2-yl)pyrimidin-2-amine | | Descriptor: | 5-fluoro-N-[5-(4-methylpiperazin-1-yl)pyridin-2-yl]-4-[(4R)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine, Cyclin-dependent kinase 6 | | Authors: | Murray, J.M. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
4LWU
 
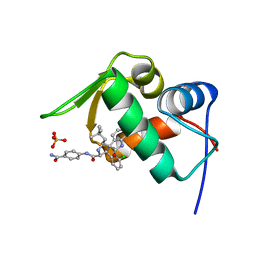 | | The 1.14A Crystal Structure of Humanized Xenopus MDM2 with RO5499252 | | Descriptor: | (2'S,3R,4'S,5'R)-N-(4-carbamoylphenyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C, Janson, C.A. | | Deposit date: | 2013-07-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Discovery of potent and selective spiroindolinone MDM2 inhibitor, RO8994, for cancer therapy.
Bioorg.Med.Chem., 22, 2014
|
|
3W2A
 
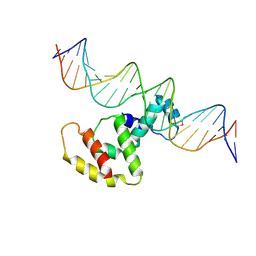 | | Crystal structure of VirB core domain complexed with the cis-acting site upstream icsp promoter | | Descriptor: | DNA (31-mer), Virulence regulon transcriptional activator VirB | | Authors: | Gao, X.P, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2012-11-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.775 Å) | | Cite: | Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes
Nucleic Acids Res., 41, 2013
|
|
3W3C
 
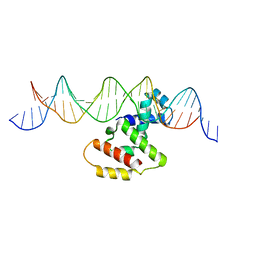 | | Crystal structure of VirB core domain complexed with the cis-acting site upstream icsb promoter | | Descriptor: | DNA (26-MER), Virulence regulon transcriptional activator VirB | | Authors: | Gao, X.P, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2012-12-19 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.431 Å) | | Cite: | Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes
Nucleic Acids Res., 41, 2013
|
|
7B0L
 
 | |
