6AEL
 
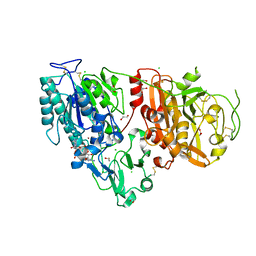 | | Crystal structure of ENPP1 in complex with 3'3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, ... | | Authors: | Kato, K, Nishimasu, H, Hirano, S, Hirano, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-08-05 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1.
Nat Commun, 9, 2018
|
|
5XH6
 
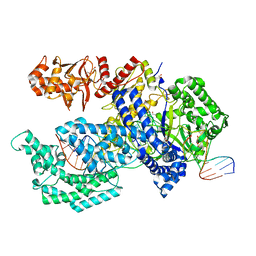 | | Crystal structure of the Acidaminococcus sp. BV3L6 Cpf1 RVR variant in complex with crRNA and target DNA (TATA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CRISPR-associated endonuclease Cpf1, ... | | Authors: | Nishimasu, H, Yamano, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1
Mol. Cell, 67, 2017
|
|
5XUS
 
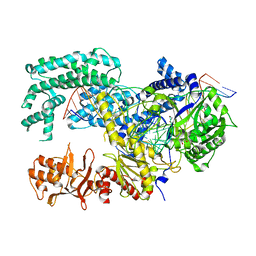 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (TTTA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*TP*TP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
7JW3
 
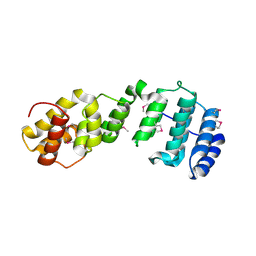 | | Crystal structure of Aedes aegypti Nibbler NTD domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7JW6
 
 | | Crystal structure of Drosophila Nibbler EXO domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2E8A
 
 | | Crystal structure of the human Hsp70 ATPase domain in complex with AMP-PNP | | Descriptor: | Heat shock 70kDa protein 1A, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Shida, M, Ishii, R, Takagi, T, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-19 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Direct inter-subdomain interactions switch between the closed and open forms of the Hsp70 nucleotide-binding domain in the nucleotide-free state.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1WPX
 
 | | Crystal structure of carboxypeptidase Y inhibitor complexed with the cognate proteinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carboxypeptidase Y, Carboxypeptidase Y inhibitor, ... | | Authors: | Mima, J, Hayashida, M, Fujii, T, Narita, Y, Hayashi, R, Ueda, M, Hata, Y. | | Deposit date: | 2004-09-14 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the carboxypeptidase y inhibitor i(c) in complex with the cognate proteinase reveals a novel mode of the proteinase-protein inhibitor interaction
J.Mol.Biol., 346, 2005
|
|
2E88
 
 | | Crystal structure of the human Hsp70 ATPase domain in the apo form | | Descriptor: | Heat shock 70kDa protein 1A, ZINC ION | | Authors: | Shida, M, Ishii, R, Takagi, T, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-19 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct inter-subdomain interactions switch between the closed and open forms of the Hsp70 nucleotide-binding domain in the nucleotide-free state.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1WJX
 
 | | Crystal sturucture of TT0801 from Thermus thermophilus | | Descriptor: | POTASSIUM ION, SsrA-binding protein | | Authors: | Bessho, Y, Shibata, R, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1WWR
 
 | | Crystal structure of tRNA adenosine deaminase TadA from Aquifex aeolicus | | Descriptor: | ZINC ION, tRNA adenosine deaminase TadA | | Authors: | Kuratani, M, Ishii, R, Bessho, Y, Fukunaga, R, Sengoku, T, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of tRNA Adenosine Deaminase (TadA) from Aquifex aeolicus
J.Biol.Chem., 280, 2005
|
|
1UFA
 
 | | Crystal structure of TT1467 from Thermus thermophilus HB8 | | Descriptor: | TT1467 protein | | Authors: | Idaka, M, Terada, T, Murayama, K, Yamaguchi, H, Nureki, O, Ishitani, R, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TT1467 from Thermus thermophilus HB8
To be published
|
|
6AID
 
 | | Structural insights into the unique polylactate degrading mechanism of Thermobifida alba cutinase | | Descriptor: | CALCIUM ION, Esterase, LACTIC ACID, ... | | Authors: | Kitadokoro, K, Kakara, M, Matsui, S, Osokoshi, R, Thumarat, U, Kawai, F, Kamitani, S. | | Deposit date: | 2018-08-22 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insights into the unique polylactate-degrading mechanism of Thermobifida alba cutinase.
Febs J., 286, 2019
|
|
5ZL6
 
 | | Histidine Racemase from Leuconostoc mesenteroides subsp. sake NBRC 102480 | | Descriptor: | Histidine racemase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Adachi, M, Shimizu, R, Oikawa, T. | | Deposit date: | 2018-03-27 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The first identification and characterization of a histidine-specific amino acid racemase, histidine racemase from a lactic acid bacterium, Leuconostoc mesenteroides subsp. sake NBRC 102480.
Amino Acids, 51, 2019
|
|
2EL7
 
 | | Crystal structure of Tryptophanyl-tRNA synthetase from Thermus thermophilus | | Descriptor: | PHOSPHATE ION, Tryptophanyl-tRNA synthetase | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Tryptophanyl-tRNA synthetase from Thermus thermophilus
To be published
|
|
2JYG
 
 | |
2JYL
 
 | |
2D5B
 
 | | Crystal Structure of Thermus Thermophilus Methionyl tRNA synthetase Y225F Mutant obtained in the presence of PEG6000 | | Descriptor: | Methionyl-tRNA Synthetase, ZINC ION | | Authors: | Konno, M, Takeda, R, Takasaka, R, Mori, Y, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-31 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Y225F/A Mutation for Met-tRNA synthetase reveals importance of hydrophobic circumstances
To be Published
|
|
2DRD
 
 | | Crystal structure of a multidrug transporter reveal a functionally rotating mechanism | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, ACRB | | Authors: | Murakami, S, Nakashima, R, Yamashita, E, Matsumoto, T. | | Deposit date: | 2006-06-08 | | Release date: | 2006-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of a multidrug transporter reveal a functionally rotating mechanism
Nature, 443, 2006
|
|
2DR6
 
 | | Crystal structure of a multidrug transporter reveal a functionally rotating mechanism | | Descriptor: | ACRB, DOXORUBICIN | | Authors: | Murakami, S, Nakashima, R, Yamashita, E, Matsumoto, T. | | Deposit date: | 2006-06-08 | | Release date: | 2006-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structures of a multidrug transporter reveal a functionally rotating mechanism
Nature, 443, 2006
|
|
2DHH
 
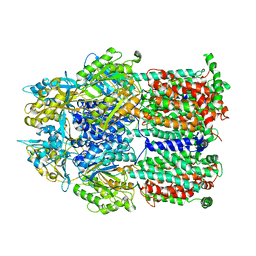 | |
2E3C
 
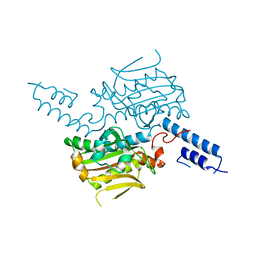 | |
3UG9
 
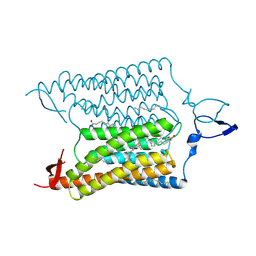 | | Crystal Structure of the Closed State of Channelrhodopsin | | Descriptor: | Archaeal-type opsin 1, Archaeal-type opsin 2, OLEIC ACID, ... | | Authors: | Kato, H.E, Ishitani, R, Nureki, O. | | Deposit date: | 2011-11-02 | | Release date: | 2012-01-25 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the channelrhodopsin light-gated cation channel
Nature, 482, 2012
|
|
3VVZ
 
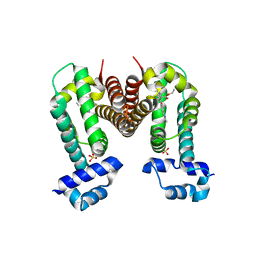 | | Crystal Strucuture of The Rhodamine 6G-Bound Form of RamR (Transcriptional Regurator of TetR Family) from Salmonella Typhimurium | | Descriptor: | Putative regulatory protein, RHODAMINE 6G, SULFATE ION | | Authors: | Sakurai, K, Nikaido, E, Nakashima, R, Yamasaki, S, Yamaguchi, A, Nishino, K. | | Deposit date: | 2012-07-30 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The crystal structure of multidrug-resistance regulator RamR with multiple drugs
Nat Commun, 4, 2013
|
|
3VVY
 
 | | Crystal Strucuture of The Ethidium-Bound Form of RamR (Transcriptional Regurator of TetR Family) from Salmonella Typhimurium | | Descriptor: | ETHIDIUM, Putative regulatory protein | | Authors: | Sakurai, K, Nikaido, E, Nakashima, R, Yamasaki, S, Yamaguchi, A, Nishino, K. | | Deposit date: | 2012-07-30 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The crystal structure of multidrug-resistance regulator RamR with multiple drugs
Nat Commun, 4, 2013
|
|
3VVX
 
 | | Crystal Strucuture of RamR (Transcriptional Regurator of TetR Family) from Salmonella Typhimurium | | Descriptor: | Putative regulatory protein, SULFATE ION | | Authors: | Sakurai, K, Nikaido, E, Nakashima, R, Yamasaki, S, Yamaguchi, A, Nishino, K. | | Deposit date: | 2012-07-30 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structure of multidrug-resistance regulator RamR with multiple drugs
Nat Commun, 4, 2013
|
|
