7E6N
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.2 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8413 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24916625 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
6QXP
 
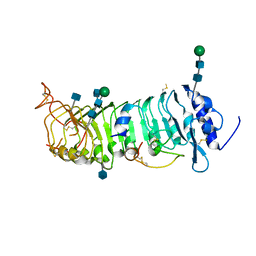 | | Protein peptide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat extensin-like protein 2, ... | | Authors: | Moussu, S, Caroline, C, Santos-Fernandez, G, Wehrle, S, Grossniklaus, U, Santiago, J. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6QWN
 
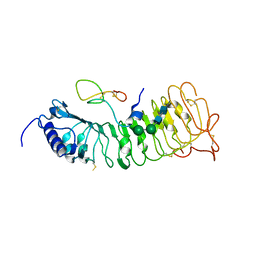 | | Protein peptide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Moussu, S, Caroline, C, Santos-Fernandez, G, Wehrle, S, Grossniklaus, U, Santiago, J. | | Deposit date: | 2019-03-05 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.892 Å) | | Cite: | Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4CLC
 
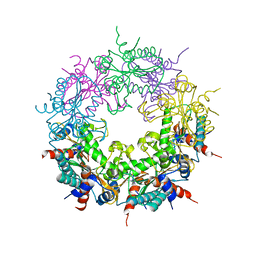 | | Crystal structure of Ybr137w protein | | Descriptor: | UPF0303 PROTEIN YBR137W | | Authors: | Yeh, Y.-H, Lin, T.-W, Lin, C.-Y, Hsiao, C.-D. | | Deposit date: | 2014-01-14 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Characterization of Ybr137Wp Implicate its Involvement in the Targeting of Tail-Anchored Proteins to Membranes.
Mol.Cell.Biol., 34, 2014
|
|
8WZB
 
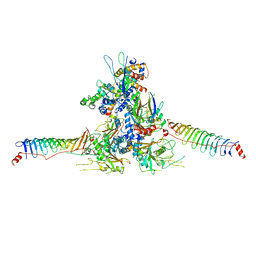 | | RS head-neck monomer | | Descriptor: | DPY30 domain containing 2, DnaJ homolog subfamily B member 13, Nucleoside diphosphate kinase homolog 5, ... | | Authors: | Meng, X, Cong, Y. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Multi-scale structures of the mammalian radial spoke and divergence of axonemal complexes in ependymal cilia.
Nat Commun, 15, 2024
|
|
8X2U
 
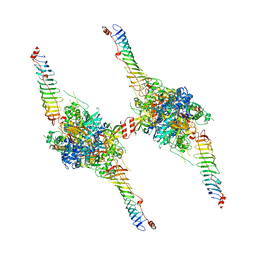 | | Radial spoke head-neck dimer | | Descriptor: | DPY30 domain containing 2, DnaJ homolog subfamily B member 13, Nucleoside diphosphate kinase homolog 5, ... | | Authors: | Meng, X, Cong, Y. | | Deposit date: | 2023-11-10 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Multi-scale structures of the mammalian radial spoke and divergence of axonemal complexes in ependymal cilia.
Nat Commun, 15, 2024
|
|
1SO8
 
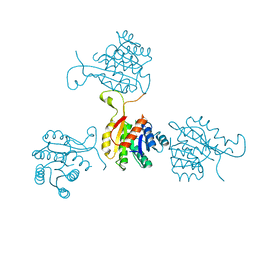 | | Abeta-bound human ABAD structure [also known as 3-hydroxyacyl-CoA dehydrogenase type II (Type II HADH), Endoplasmic reticulum-associated amyloid beta-peptide binding protein (ERAB)] | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type II, CHLORIDE ION, SODIUM ION | | Authors: | Lustbader, J.W, Cirilli, M, Wu, H. | | Deposit date: | 2004-03-13 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease.
Science, 304, 2004
|
|
1BT7
 
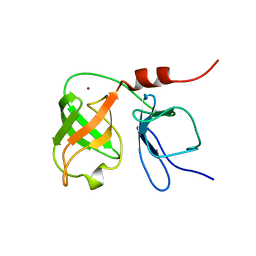 | | THE SOLUTION NMR STRUCTURE OF THE N-TERMINAL PROTEASE DOMAIN OF THE HEPATITIS C VIRUS (HCV) NS3-PROTEIN, FROM BK STRAIN, 20 STRUCTURES | | Descriptor: | NS3 SERINE PROTEASE, ZINC ION | | Authors: | Barbato, G, Cicero, D.O, Nardi, M.C, Steinkuhler, C, Cortese, R, De Francesco, R, Bazzo, R. | | Deposit date: | 1998-09-01 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism.
J.Mol.Biol., 289, 1999
|
|
3EGA
 
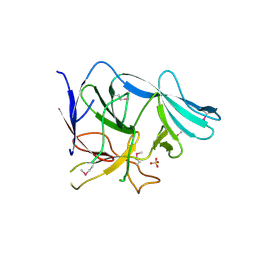 | |
3EGB
 
 | |
2GVF
 
 | | HCV NS3-4A protease domain complexed with a macrocyclic ketoamide inhibitor, SCH419021 | | Descriptor: | (6R,8S,11S)-11-CYCLOHEXYL-N-(1-{[(2-{[(1S)-2-(DIMETHYLAMINO)-2-OXO-1-PHENYLETHYL]AMINO}-2-OXOETHYL)AMINO](OXO)ACETYL}BUTYL)-10,13-DIOXO-2,5-DIOXA-9,12-DIAZATRICYCLO[14.3.1.1~6,9~]HENICOSA-1(20),16,18-TRIENE-8-CARBOXAMIDE, Polyprotein, ZINC ION, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chen, K.X, Venkatraman, S, Parekh, T.N, Gu, H, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V, Girijavallabhan, V. | | Deposit date: | 2006-05-02 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4GL2
 
 | | Structural Basis for dsRNA duplex backbone recognition by MDA5 | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RNA (5'-R(*AP*UP*CP*CP*GP*CP*GP*GP*CP*CP*CP*U)-3'), ... | | Authors: | Wu, B, Hur, S. | | Deposit date: | 2012-08-13 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.557 Å) | | Cite: | Structural Basis for dsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5.
Cell(Cambridge,Mass.), 152, 2013
|
|
2EFJ
 
 | |
2EG5
 
 | | The structure of xanthosine methyltransferase | | Descriptor: | 9-[(2R,3R,4S,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)OXOLAN-2-YL]-3H-PURINE-2,6-DIONE, S-ADENOSYL-L-HOMOCYSTEINE, Xanthosine methyltransferase | | Authors: | McCarthy, A.A, McCarthy, J.G. | | Deposit date: | 2007-02-28 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of two N-methyltransferases from the caffeine biosynthetic pathway
Plant Physiol., 144, 2007
|
|
2F9V
 
 | | HCV NS3 protease domain with NS4a peptide and a ketoamide inhibitor with P1 and P2 cyclopropylalannines | | Descriptor: | (2S,8R,9S,15S)-15-CYCLOHEXYL-9,12-BIS(CYCLOPROPYLMETHYL)-8-HYDROXY-20-METHYL-4,7,11,14,17-PENTAOXO-2-PHENYL-18-OXA-3,6,10,12,13,16-HEXAAZAHENICOSAN-1-OIC ACID, NS3 protease/helicase, ZINC ION, ... | | Authors: | Bogen, S.L, Ruan, S, Liu, R, Agrawal, S, Pichardo, J, Prongay, A, Baroudy, B, Saksena, A, Girijavallabhan, V, Njoroge, F.G. | | Deposit date: | 2005-12-06 | | Release date: | 2007-01-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Depeptidization efforts on P3-P2 a-ketoamide inhibitors of HCV NS3-4A serine protease: Effect on HCV replicon activity.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2F9U
 
 | | HCV NS3 protease domain with NS4a peptide and a ketoamide inhibitor with a P2 norborane | | Descriptor: | 1,1-DIMETHYLETHYL [1-CYCLOHEXYL-2-[3-[[[1-[2-[[2-[[2-(DIMETHYLAMINO)-2-OXO-1-PHENYLETHYL]AMINO]-2-OXOETHYL]AMINO]-1,2-DIOXOETHYL]PENTYL]AMINO]CARBONYL]-2-AZABICYCLO[2.2.1]HEPTAN-2-YL]-2-OXOETHYL]CARBAMATE, NS3 protease/helicase', ZINC ION, ... | | Authors: | Venkatraman, S, Njoroge, F.G, Wu, W, Girijavallabhan, V, Prongay, A.J, Butkiewicz, N, Pichardo, J. | | Deposit date: | 2005-12-06 | | Release date: | 2006-06-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel Inhibitors of Hepatitis C NS3-NS4A Serine Protease Derived from 2-Aza-bicyclo[2.2.1]heptane-3-carboxylic acid.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4W7J
 
 | |
4W7K
 
 | |
4W7M
 
 | |
8QEY
 
 | | Structure of human Asc1/CD98hc heteromeric amino acid transporter | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Martinez-Molledo, M, Rullo-Tubau, J, Errasti-Murugarren, E, Palacin, M, Llorca, O. | | Deposit date: | 2023-09-01 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and mechanisms of transport of human Asc1/CD98hc amino acid transporter.
Nat Commun, 15, 2024
|
|
7T0O
 
 | |
7T0R
 
 | | Crystal structure of the anti-CD4 adnectin 6940_B01 as a complex with the extracellular domains of CD4 and ibalizumab fAb | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adnectin 6940_B01, Ibalizumab Heavy Chain, ... | | Authors: | Williams, S.P, Concha, N.O, Wensel, D.L, Hong, X. | | Deposit date: | 2021-11-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Novel Bent Conformation of CD4 Induced by HIV-1 Inhibitor Indirectly Prevents Productive Viral Attachment.
J.Mol.Biol., 434, 2021
|
|
6S4A
 
 | | Structure of human MTHFD2 in complex with TH9028 | | Descriptor: | (2~{S})-2-[[5-[[2,4-bis(azanyl)-6-oxidanylidene-5~{H}-pyrimidin-5-yl]carbamoylamino]pyridin-2-yl]carbonylamino]-4-(1~{H}-1,2,3,4-tetrazol-5-yl)butanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Gustafsson, R, Scaletti, E.R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
6S4F
 
 | | Structure of human MTHFD2 in complex with TH9619 | | Descriptor: | (E,4S)-4-[[5-[2-[2,6-bis(azanyl)-4-oxidanylidene-1H-pyrimidin-5-yl]ethanoylamino]-3-fluoranyl-pyridin-2-yl]carbonylamino]pent-2-enedioic acid, ADENOSINE-5'-DIPHOSPHATE, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Scaletti, E.R, Gustafsson, R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
