2L3R
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Histone H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3 | | Authors: | Nady, N, Lemak, A, Fares, C, Gutmanas, A, Avvakumov, G, Xue, S, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-21 | | Release date: | 2011-04-13 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Recognition of Multivalent Histone States Associated with Heterochromatin by UHRF1 Protein.
J.Biol.Chem., 286, 2011
|
|
4F09
 
 | |
3T9B
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP at pH 5.2 | | Descriptor: | Inositol Pyrophosphate Kinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3TCL
 
 | | Crystal Structure of HIV-1 Neutralizing Antibody CH04 | | Descriptor: | CH04 Heavy Chain Fab, CH04 Light Chain Fab, IMIDAZOLE | | Authors: | Louder, R.K, McLellan, J.S, Pancera, M, Yang, Y, Zhang, B, Bonsignori, M, Kwong, P.D. | | Deposit date: | 2011-08-09 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3TGT
 
 | | Crystal structure of unliganded HIV-1 clade A/E strain 93TH057 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 clade A/E 93TH057 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7M0G
 
 | |
2LWE
 
 | |
4G1Q
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with Rilpivirine (TMC278, Edurant), a non-nucleoside rt-inhibiting drug | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, MAGNESIUM ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2012-07-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Snapshot of the equilibrium dynamics of a drug bound to HIV-1 reverse transcriptase.
Nat Chem, 5, 2013
|
|
4G2S
 
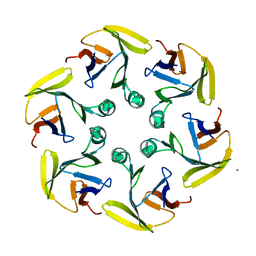 | |
4G31
 
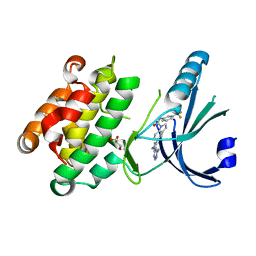 | | Crystal Structure of GSK6414 Bound to PERK (R587-R1092, delete A660-T867) at 2.28 A Resolution | | Descriptor: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-2,3-dihydro-1H-indol-1-yl]-2-[3-(trifluoromethyl)phenyl]ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3, GLYCEROL | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 55, 2012
|
|
3T7A
 
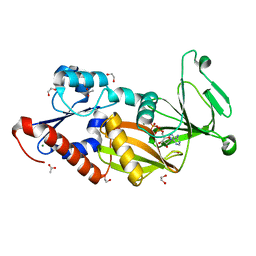 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP at pH 5.2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-07-29 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T54
 
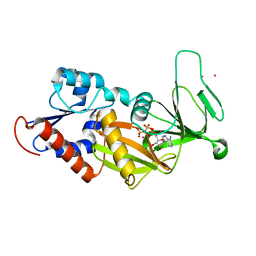 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ATP and Cadmium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CADMIUM ION, Inositol Pyrophosphate Kinase | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-07-26 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T9F
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and 1,5-(PP)2-IP4 (1,5-IP8) | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], ADENOSINE-5'-DIPHOSPHATE, CADMIUM ION, ... | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
4G9F
 
 | | Crystal Structure of C12C TCR-HLAB2705-KK10-L6M | | Descriptor: | Beta-2-microglobulin, Gag protein, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Wilmann, P.G, Rossjohn, J. | | Deposit date: | 2012-07-23 | | Release date: | 2013-03-20 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Molecular Basis for the Control of Preimmune Escape Variants by HIV-Specific CD8(+) T Cells.
Immunity, 38, 2013
|
|
2LWD
 
 | |
7NO8
 
 | | Structure of the mature RSV CA lattice: Group I, pentamer-hexamer interface, class 1"6 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOM
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 4'Beta | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
4FXJ
 
 | |
7NO6
 
 | | Structure of the mature RSV CA lattice: Group I, pentamer-hexamer interface, class 1 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOG
 
 | | Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 4'5 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO0
 
 | | Structure of the mature RSV CA lattice: T=1 CA icosahedron | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOB
 
 | | Structure of the mature RSV CA lattice: Group II, hexamer-hexamer interface, class 2'6 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NON
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 4'Gamma | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO1
 
 | | Structure of the mature RSV CA lattice: T=3 CA icosahedron | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOE
 
 | | Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 3'5 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
