7DQA
 
 | | Cryo-EM structure of SARS-CoV2 RBD-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Wang, J, Lan, J, Wang, X.Q, Wang, H.W. | | Deposit date: | 2020-12-22 | | Release date: | 2021-12-29 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Reduced graphene oxide membrane as supporting film for high-resolution cryo-EM
Biophys Rep, 7, 2022
|
|
224L
 
 | |
206L
 
 | | PHAGE T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1996-03-19 | | Release date: | 1996-08-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Energetic cost and structural consequences of burying a hydroxyl group within the core of a protein determined from Ala-->Ser and Val-->Thr substitutions in T4 lysozyme.
Biochemistry, 32, 1993
|
|
6BZ1
 
 | | MEF2 Chimera D83V mutant/DNA complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*A)-3'), DNA (5'-D(P*TP*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*TP*T)-3'), MEF2 CHIMERA | | Authors: | Lei, X, Chen, L. | | Deposit date: | 2017-12-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | The Cancer Mutation D83V Induces an alpha-Helix to beta-Strand Conformation Switch in MEF2B.
J. Mol. Biol., 430, 2018
|
|
6BYY
 
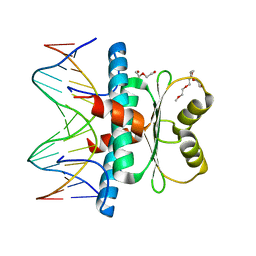 | | MEF2 CHIMERA/DNA Complex | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 3,3',3''-phosphanetriyltripropanoic acid, DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*A)-3'), ... | | Authors: | Lei, X, Chen, L. | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Cancer Mutation D83V Induces an alpha-Helix to beta-Strand Conformation Switch in MEF2B.
J. Mol. Biol., 430, 2018
|
|
6CKS
 
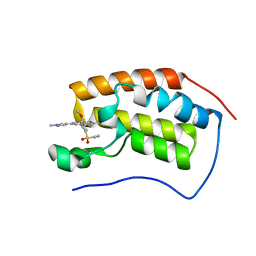 | | Crystal Structure of BRD4 with QC4956 | | Descriptor: | 4-[5-(ethylsulfonyl)-2-methoxyphenyl]-2-methyl-6-(1-methyl-1H-pyrazol-4-yl)isoquinolin-1(2H)-one, Bromodomain-containing protein 4 | | Authors: | Hosfield, D.J. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Design, synthesis and biological evaluation of novel 4-phenylisoquinolinone BET bromodomain inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6CKR
 
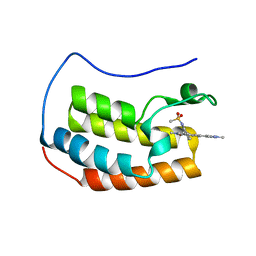 | | Crystal Structure of BRD4 with QC4956 | | Descriptor: | Bromodomain-containing protein 4, N-{3-[2-methyl-6-(1-methyl-1H-pyrazol-4-yl)-1-oxo-1,2-dihydroisoquinolin-4-yl]phenyl}methanesulfonamide | | Authors: | Hosfield, D.J. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Design, synthesis and biological evaluation of novel 4-phenylisoquinolinone BET bromodomain inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6K41
 
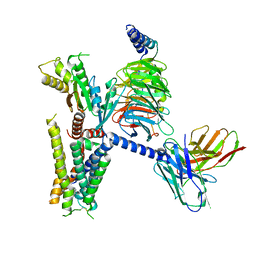 | | cryo-EM structure of alpha2BAR-GoA complex | | Descriptor: | 4-[(1~{S})-1-(2,3-dimethylphenyl)ethyl]-1~{H}-imidazole, Alpha-2A adrenergic receptor,Endolysin,Alpha-2B adrenergic receptor,Alpha-2B adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, D, Liu, Z, Wang, H.W, Kobilka, B.K. | | Deposit date: | 2019-05-23 | | Release date: | 2020-04-15 | | Last modified: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation of the alpha2Badrenoceptor by the sedative sympatholytic dexmedetomidine.
Nat.Chem.Biol., 16, 2020
|
|
6K42
 
 | | cryo-EM structure of alpha2BAR-Gi1 complex | | Descriptor: | 4-[(1~{S})-1-(2,3-dimethylphenyl)ethyl]-1~{H}-imidazole, Alpha-2A adrenergic receptor,Endolysin,Alpha-2B adrenergic receptor,Alpha-2B adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, D, Liu, Z, Wang, H.W, Kobilka, B.K. | | Deposit date: | 2019-05-23 | | Release date: | 2020-04-15 | | Last modified: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Activation of the alpha2Badrenoceptor by the sedative sympatholytic dexmedetomidine.
Nat.Chem.Biol., 16, 2020
|
|
8K4F
 
 | | DHODH in complex with compound A0 | | Descriptor: | 5-cyclopropyl-2-[1-[(2-fluorophenyl)methyl]pyrazolo[3,4-b]pyridin-3-yl]pyrimidin-4-amine, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, ACETATE ION, ... | | Authors: | Jian, L, Sun, Q. | | Deposit date: | 2023-07-18 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery and Optimization of Novel h DHODH Inhibitors for the Treatment of Inflammatory Bowel Disease.
J.Med.Chem., 66, 2023
|
|
3QWX
 
 | | CED-2 1-174 | | Descriptor: | Cell death abnormality protein 2, SULFATE ION | | Authors: | Kang, Y, Sun, J, Liu, Y. | | Deposit date: | 2011-02-28 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the cell corpse engulfment protein CED-2 in Caenorhabditis elegans.
Biochem.Biophys.Res.Commun., 410, 2011
|
|
3QWY
 
 | | CED-2 | | Descriptor: | Cell death abnormality protein 2, GLYCEROL, SULFATE ION | | Authors: | Kang, Y, Sun, J, Liu, Y, Sun, D, Hu, Y, Liu, Y.F. | | Deposit date: | 2011-02-28 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of the cell corpse engulfment protein CED-2 in Caenorhabditis elegans.
Biochem.Biophys.Res.Commun., 410, 2011
|
|
8GIA
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) Nsp3 macrodomain in complex with TFMU-ADPr | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl [(2R,3S,4R,5R)-3,4-dihydroxy-5-{[2-oxo-4-(trifluoromethyl)-2H-1-benzopyran-7-yl]oxy}oxolan-2-yl]methyl dihydrogen diphosphate | | Authors: | Wallace, S.D, Bagde, S.R, Fromme, J.C. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Fluorescence Polarization Assay for Macrodomains Facilitates the Identification of Potent Inhibitors of the SARS-CoV-2 Macrodomain.
Acs Chem.Biol., 18, 2023
|
|
6KNK
 
 | | Crystal structure of SbnH in complex with citryl-diaminoethane | | Descriptor: | (2S)-2-{2-[(2-AMINOETHYL)AMINO]-2-OXOETHYL}-2-HYDROXYBUTANEDIOIC ACID, (2~{S})-2-[2-[2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]ethylamino]-2-oxidanylidene-ethyl]-2-oxidanyl-butanedioic acid, PHOSPHATE ION, ... | | Authors: | Tang, J, Ju, Y, Zhou, H. | | Deposit date: | 2019-08-05 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into Substrate Recognition and Activity Regulation of the Key Decarboxylase SbnH in Staphyloferrin B Biosynthesis.
J.Mol.Biol., 431, 2019
|
|
6KNI
 
 | |
6KNH
 
 | | Crystal structure of SbnH in complex with citrate, a PLP-dependent decarboxylase in Staphyloferrin B biothesynthesis | | Descriptor: | CITRIC ACID, PHOSPHATE ION, Probable diaminopimelate decarboxylase protein | | Authors: | Tang, J, Ju, Y, Zhou, H. | | Deposit date: | 2019-08-05 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into Substrate Recognition and Activity Regulation of the Key Decarboxylase SbnH in Staphyloferrin B Biosynthesis.
J.Mol.Biol., 431, 2019
|
|
1VHN
 
 | |
119L
 
 | |
118L
 
 | |
120L
 
 | |
123L
 
 | |
125L
 
 | |
126L
 
 | |
128L
 
 | |
127L
 
 | |
