7CI4
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (2R)-2-azanyl-4-methylsulfonyl-N-[3-(trifluoromethyloxy)phenyl]butanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CIC
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 2-azanyl-N-[3-(trifluoromethyloxy)phenyl]ethanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI9
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-2-[[4-[2-[3-[[2-[(1S)-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]ethynyl]phenoxy]methyl]propane-1,3-diol, PHOSPHATE ION, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI5
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (3R)-3-azanyl-4-oxidanylidene-4-[[3-(trifluoromethyloxy)phenyl]amino]butanoic acid, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
2D8B
 
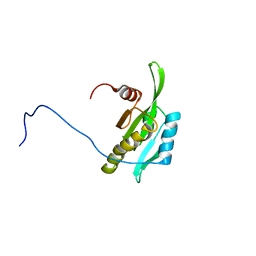 | | Solution structure of the second tandem cofilin-domain of mouse twinfilin | | Descriptor: | Twinfilin-1 | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Sato, M, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-02 | | Release date: | 2006-06-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains
Protein Sci., 18, 2009
|
|
2DWV
 
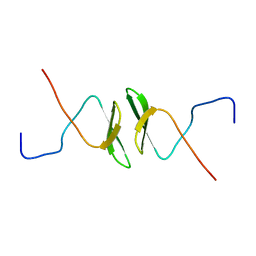 | | Solution structure of the second WW domain from mouse salvador homolog 1 protein (mWW45) | | Descriptor: | Salvador homolog 1 protein | | Authors: | Ohnishi, S, Kigawa, T, Koshiba, S, Tomizawa, T, Sato, M, Tochio, N, Inoue, M, Harada, T, Watanabe, S, Guntert, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-17 | | Release date: | 2007-02-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an atypical WW domain in a novel beta-clam-like dimeric form
Febs Lett., 581, 2007
|
|
2CXF
 
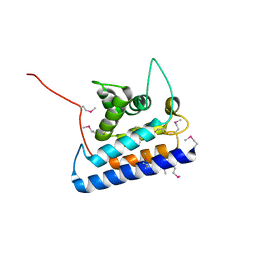 | | RUN domain of Rap2 interacting protein x, crystallized in C2 space group | | Descriptor: | rap2 interacting protein x | | Authors: | Kukimoto-Niino, M, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-29 | | Release date: | 2005-12-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Crystal Structure of the RUN Domain of the RAP2-interacting Protein x
J.Biol.Chem., 281, 2006
|
|
2CXL
 
 | | RUN domain of Rap2 interacting protein x, crystallized in I422 space group | | Descriptor: | rap2 interacting protein x | | Authors: | Kukimoto-Niino, M, Umehara, T, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-30 | | Release date: | 2005-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the RUN Domain of the RAP2-interacting Protein x
J.Biol.Chem., 281, 2006
|
|
2DWG
 
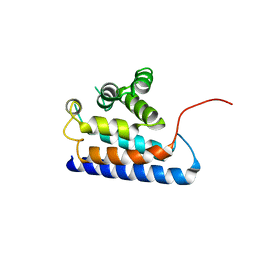 | | RUN domain of Rap2 interacting protein x, crystallized in P2(1)2(1)2(1) space group | | Descriptor: | Protein RUFY3 | | Authors: | Kukimoto-Niino, M, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-11 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Crystal Structure of the RUN Domain of the RAP2-interacting Protein x
J.Biol.Chem., 281, 2006
|
|
2DWK
 
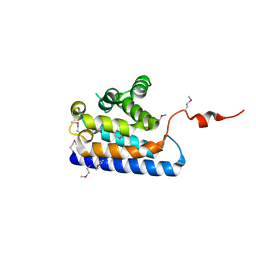 | | Crystal structure of the RUN domain of mouse Rap2 interacting protein x | | Descriptor: | Protein RUFY3 | | Authors: | Kukimoto-Niino, M, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-15 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the RUN Domain of the RAP2-interacting Protein x
J.Biol.Chem., 281, 2006
|
|
2E5S
 
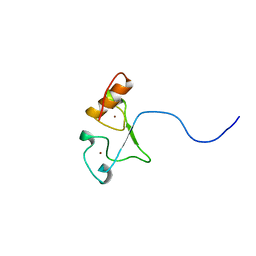 | | Solution structure of the zf-CCCHx2 domain of muscleblind-like 2, isoform 1 [Homo sapiens] | | Descriptor: | OTTHUMP00000018578, ZINC ION | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-22 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain in the human muscleblind-like protein 2
Protein Sci., 18, 2009
|
|
7W3L
 
 | | Crystal structure of LSD1 in complex with cis-4-Br-2,5-F2-PCPA (S1024) | | Descriptor: | 3-[4-bromanyl-2,5-bis(fluoranyl)phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
7XE1
 
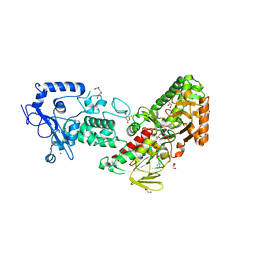 | | Crystal structure of LSD2 in complex with cis-4-Br-PCPA | | Descriptor: | 3-(4-bromophenyl)propanal, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
7XE2
 
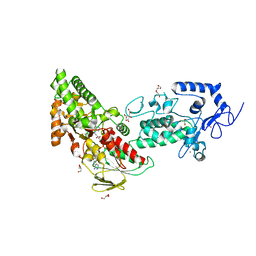 | | Crystal structure of LSD2 in complex with trans-4-Br-PCPA | | Descriptor: | 3-(4-bromophenyl)propanal, CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
7XE3
 
 | | Crystal structure of LSD2 in complex with cis-4-Br-2,5-F2-PCPA (S1024) | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-bromanyl-2,5-bis(fluoranyl)phenyl]propanal, CITRATE ANION, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
7VQT
 
 | | Crystal structure of LSD1 in complex with compound 5 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-4-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
7VQS
 
 | | Crystal structure of LSD1 in complex with compound 4 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-3-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
7VQU
 
 | | Crystal structure of LSD1 in complex with compound S1427 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-3-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
6NZV
 
 | | Crystal structure of HCV NS3/4A protease in complex with compound 12 | | Descriptor: | (1aR,5S,8S,9S,10R,22aR)-5-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-9-ethyl-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, HCV NS3/4A protease, SULFATE ION, ... | | Authors: | Appleby, T.C, Taylor, J.G. | | Deposit date: | 2019-02-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of the pan-genotypic hepatitis C virus NS3/4A protease inhibitor voxilaprevir (GS-9857): A component of Vosevi®.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3VS1
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 1-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]-3-phenylurea | | Descriptor: | 1-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]-3-phenylurea, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Toyama, M, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.464 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS4
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 5-(4-phenoxyphenyl)-7-(tetrahydro-2H-pyran-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 5-(4-phenoxyphenyl)-7-(tetrahydro-2H-pyran-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.747 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS0
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor N-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]benzamide | | Descriptor: | CALCIUM ION, CHLORIDE ION, N-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]benzamide, ... | | Authors: | Kuratani, M, Honda, K, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.934 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS3
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomaebchi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS6
 
 | | Crystal structure of HCK complexed with a pyrazolo-pyrimidine inhibitor tert-butyl {4-[4-amino-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl]-2-methoxyphenyl}carbamate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Tyrosine-protein kinase HCK, ... | | Authors: | Kuratani, M, Honda, K, Tomabechi, Y, Toyama, M, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.373 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS5
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-(1-methylpiperidin-4-yl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-(1-methylpiperidin-4-yl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, Tyrosine-protein kinase HCK | | Authors: | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
