8SBC
 
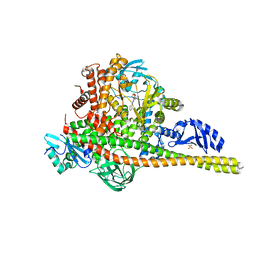 | | Co-structure of Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform and brain penetrant inhibitors | | Descriptor: | (2M)-7-[(3R)-3-methylmorpholin-4-yl]-5-[(3S)-3-methylmorpholin-4-yl]-2-(pyridin-2-yl)-1H-imidazo[4,5-b]pyridine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Knapp, M.S, Elling, R.A, Tang, J. | | Deposit date: | 2023-04-03 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Brain-Penetrant ATP-Competitive mTOR Inhibitors for CNS Syndromes.
J.Med.Chem., 66, 2023
|
|
7C7E
 
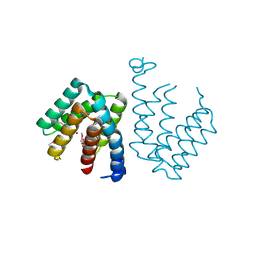 | | Crystal structure of C terminal domain of Escherichia coli DgoR | | Descriptor: | Putative DNA-binding transcriptional regulator, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Lin, W. | | Deposit date: | 2020-05-25 | | Release date: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural and Functional Analyses of the Transcription Repressor DgoR From Escherichia coli Reveal a Divalent Metal-Containing D-Galactonate Binding Pocket.
Front Microbiol, 11, 2020
|
|
7CWS
 
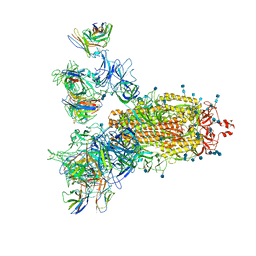 | | SARS-CoV-2 Spike Proteins Trimer in Complex with FC05 and H014 Fabs Cocktail | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of FC05 Fab, ... | | Authors: | Wang, L, Wang, X. | | Deposit date: | 2020-08-31 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based development of human antibody cocktails against SARS-CoV-2.
Cell Res., 31, 2021
|
|
7CWU
 
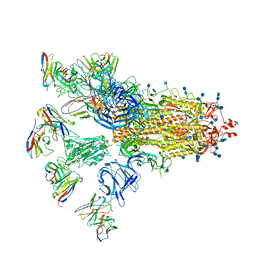 | | SARS-CoV-2 spike proteins trimer in complex with P17 and FC05 Fabs cocktail | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-31 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-based development of human antibody cocktails against SARS-CoV-2.
Cell Res., 31, 2021
|
|
7CWT
 
 | |
7D3B
 
 | | flavone reductase | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, Cd1, FLAVIN MONONUCLEOTIDE | | Authors: | Hong, S, Yang, G.H, Zhang, P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
7D3A
 
 | | flavone reductase | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Cd1, FLAVIN MONONUCLEOTIDE | | Authors: | Hong, S, Yang, G.H, Zhang, P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-03 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
7D39
 
 | | FLR-apo | | Descriptor: | Cd1, FLAVIN MONONUCLEOTIDE | | Authors: | Hong, S, Yang, G.H, Zhang, P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
7D38
 
 | | flavone reductase | | Descriptor: | Cd1, FLAVIN MONONUCLEOTIDE, chrysin | | Authors: | Hong, S, Yang, G.H, Zhang, P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
7DST
 
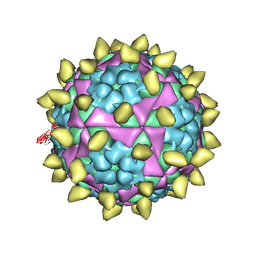 | | FMDV capsid in complex with M170 Nab | | Descriptor: | M170 Nab, VP1 of O type FMDV capsid, VP2 of O type FMDV capsid, ... | | Authors: | Dong, H, Liu, P. | | Deposit date: | 2021-01-02 | | Release date: | 2021-03-10 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and molecular basis for foot-and-mouth disease virus neutralization by two potent protective antibodies.
Protein Cell, 13, 2022
|
|
7DSS
 
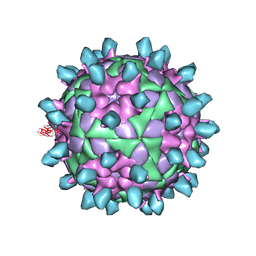 | | Complex of FMDV and M8 Nab | | Descriptor: | M8 Nab, VP1 of O type FMDV capsid, VP2 of O-type FMDV capsid, ... | | Authors: | Dong, H, Liu, P. | | Deposit date: | 2021-01-02 | | Release date: | 2021-03-31 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and molecular basis for foot-and-mouth disease virus neutralization by two potent protective antibodies.
Protein Cell, 13, 2022
|
|
7E7B
 
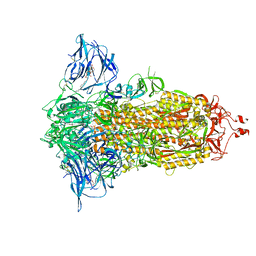 | | Cryo-EM structure of the SARS-CoV-2 furin site mutant S-Trimer from a subunit vaccine candidate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside, ... | | Authors: | Zheng, S, Ma, J. | | Deposit date: | 2021-02-25 | | Release date: | 2021-03-24 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of S-Trimer, a subunit vaccine candidate for COVID-19.
J.Virol., 95, 2021
|
|
7E7D
 
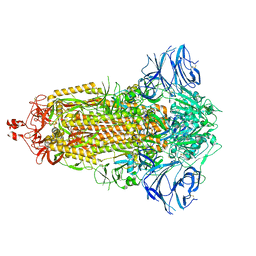 | |
7EG4
 
 | | Cryo-EM structure of nauclefine-induced PDE3A-SLFN12 complex | | Descriptor: | MAGNESIUM ION, Parvine, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
7EG0
 
 | | Cryo-EM structure of anagrelide-induced PDE3A-SLFN12 complex | | Descriptor: | 6,7-bis(chloranyl)-3,5-dihydro-1H-imidazo[2,1-b]quinazolin-2-one, MAGNESIUM ION, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-23 | | Release date: | 2021-09-29 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
7EG1
 
 | | Cryo-EM structure of DNMDP-induced PDE3A-SLFN12 complex | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-23 | | Release date: | 2021-11-03 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
6L4S
 
 | |
7WRW
 
 | | Structure of Deinococcus radiodurans HerA | | Descriptor: | HerA | | Authors: | Cheng, K. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.00008273 Å) | | Cite: | Structural and DNA end resection study of the bacterial NurA-HerA complex.
Bmc Biol., 21, 2023
|
|
7WRX
 
 | | Structure of Deinococcus radiodurans HerA-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HerA, MAGNESIUM ION | | Authors: | Cheng, K. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.40003562 Å) | | Cite: | Structural and DNA end resection study of the bacterial NurA-HerA complex.
Bmc Biol., 21, 2023
|
|
7XJP
 
 | | Cryo-EM structure of EDS1 and SAG101 with ATP-APDR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5-DIPHOSPHORIBOSE, ISOPROPYL ALCOHOL, ... | | Authors: | Huang, S.J, Jia, A.L, Han, Z.F, Chai, J.J. | | Deposit date: | 2022-04-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity.
Science, 377, 2022
|
|
7XNA
 
 | | Crystal structure of somatostatin receptor 2 (SSTR2) with peptide antagonist CYN 154806 | | Descriptor: | CYN 154806, Somatostatin receptor type 2,Endo-1,4-beta-xylanase | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XN9
 
 | | Crystal structure of SSTR2 and L-054,522 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Somatostatin receptor type 2,Endo-1,4-beta-xylanase, tert-butyl (2S)-6-azanyl-2-[[(2R,3S)-3-(1H-indol-3-yl)-2-[[4-(2-oxidanylidene-3H-benzimidazol-1-yl)piperidin-1-yl]carbonylamino]butanoyl]amino]hexanoate | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
5X62
 
 | |
5YAC
 
 | | Crystal structure of WT Trm5b from Pyrococcus abyssi | | Descriptor: | tRNA (guanine(37)-N1)-methyltransferase Trm5b | | Authors: | Xie, W, Wu, J. | | Deposit date: | 2017-08-31 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The crystal structure of the Pyrococcus abyssi mono-functional methyltransferase PaTrm5b
Biochem. Biophys. Res. Commun., 493, 2017
|
|
6KVL
 
 | | Crystal structure of UDP-RebB-SrUGT76G1 | | Descriptor: | (8alpha,9beta,10alpha,13alpha)-13-{[beta-D-glucopyranosyl-(1->2)-[beta-D-glucopyranosyl-(1->3)]-beta-D-glucopyranosyl]oxy}kaur-16-en-18-oic acid, UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
