8ILS
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 17 | | Descriptor: | N-[(2R)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ILR
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 16 | | Descriptor: | N-[(2S)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
9LK5
 
 | | The structure of Lhcb8-C2S2 PSII-LHCII supercomplex from Arabidopsis thaliana | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Zhou, Q, Caferri, R, Shan, J.Y, Amelii, A, Bassi, R, Liu, Z.F. | | Deposit date: | 2025-01-16 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A stress-induced paralog of Lhcb4 controls the photosystem II functional architecture in Arabidopsis thaliana
To Be Published
|
|
9LK4
 
 | | Cryo-EM structure of Lhcb4.1-C2S2 PSII-LHCII supercomplex from Arabidopsis thaliana | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Zhou, Q, Caferri, R, Shan, J.Y, Amelii, A, Bassi, R, Liu, Z.F. | | Deposit date: | 2025-01-16 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A stress-induced paralog of Lhcb4 controls the photosystem II functional architecture in Arabidopsis thaliana
To Be Published
|
|
6WVW
 
 | | Crystal structure of the R59P-SNAP25 containing SNARE complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Synaptosomal-associated protein 25, ... | | Authors: | Zhou, Q, White, K.I, Brunger, A.T. | | Deposit date: | 2020-05-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Role of Aberrant Spontaneous Neurotransmission in SNAP25-Associated Encephalopathies.
Neuron, 109, 2021
|
|
1TYZ
 
 | | Crystal structure of 1-Aminocyclopropane-1-carboyxlate Deaminase from Pseudomonas | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-08 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes:Insight into the mechanism of unique pyrodoxial-5'-phosphate dependent cyclopropane ring opening reaction
Biochemistry, 43, 2004
|
|
1TZK
 
 | | Crystal structure of 1-aminocyclopropane-1-carboxylate-deaminase complexed with alpha-keto-butyrate | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, 2-KETOBUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-10 | | Release date: | 2004-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
1TZM
 
 | | Crystal structure of ACC deaminase complexed with substrate analog b-chloro-D-alanine | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, 3-chloro-D-alanine, AMINO-ACRYLATE, ... | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-10 | | Release date: | 2004-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
1TZJ
 
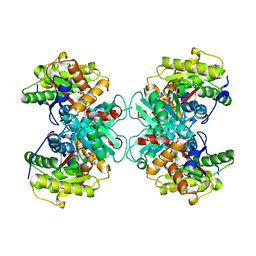 | | Crystal Structure of 1-aminocyclopropane-1-carboxylate deaminase complexed with d-vinyl glycine | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, D-VINYLGLYCINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-10 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
1TZ2
 
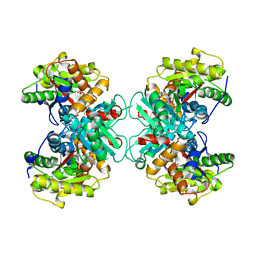 | | Crystal structure of 1-aminocyclopropane-1-carboyxlate deaminase complexed with ACC | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-09 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
3JUE
 
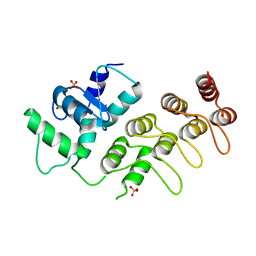 | | Crystal Structure of ArfGAP and ANK repeat domain of ACAP1 | | Descriptor: | ARFGAP with coiled-coil, ANK repeat and PH domain-containing protein 1, SULFATE ION, ... | | Authors: | Pang, X, Zhang, K, Ma, J, Zhou, Q, Sun, F. | | Deposit date: | 2009-09-15 | | Release date: | 2010-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
2B34
 
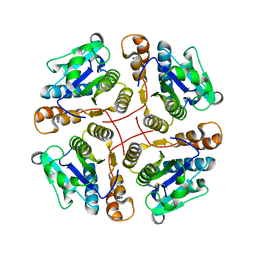 | | Structure of MAR1 Ribonuclease from Caenorhabditis elegans | | Descriptor: | MAR1 Ribonuclease | | Authors: | Schormann, N, Karpova, E, Li, S, Symersky, J, Zhang, Y, Lu, S, Zhou, Q, Lin, G, Cao, Z, Luo, M, Qiu, S, Luan, C.-H, Luo, D, Huang, W, Shang, Q, McKinstry, A, An, J, Tsao, J, Carson, M, Stinnett, M, Chen, Y, Johnson, D, Gary, R, Arabshahi, A, Bunzel, R, Bray, T, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-09-19 | | Release date: | 2005-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structure of MAR1 Ribonuclease from Caenorhabditis elegans
To be Published
|
|
3A2A
 
 | | The structure of the carboxyl-terminal domain of the human voltage-gated proton channel Hv1 | | Descriptor: | CHLORIDE ION, Voltage-gated hydrogen channel 1 | | Authors: | Li, S.J, Unno, H, Zhou, Q, Zhao, Q, Zhai, Y, Sun, F. | | Deposit date: | 2009-05-08 | | Release date: | 2010-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role and structure of the carboxyl-terminal domain of the human voltage-gated proton channel Hv1.
J.Biol.Chem., 285, 2010
|
|
3JD8
 
 | | cryo-EM structure of the full-length human NPC1 at 4.4 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Gong, X, Qian, H.W, Zhou, X.H, Wu, J.P, Zhou, Q, Yan, N. | | Deposit date: | 2016-05-01 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | The structure of Niemann-Pick C1 protein
To Be Published
|
|
5KJ7
 
 | | Structure of the Ca2+-bound synaptotagmin-1 SNARE complex (long unit cell form) - from XFEL diffraction | | Descriptor: | CALCIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Lyubimov, A.Y, Uervirojnangkoorn, M, Zhou, Q, Zhao, M, Sauter, N.K, Brewster, A.S, Weis, W.I, Brunger, A.T. | | Deposit date: | 2016-06-17 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Advances in X-ray free electron laser (XFEL) diffraction data processing applied to the crystal structure of the synaptotagmin-1 / SNARE complex.
Elife, 5, 2016
|
|
5KJ8
 
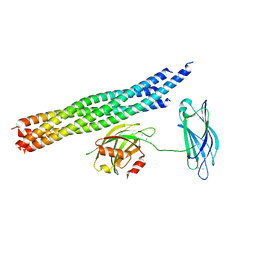 | | Structure of the Ca2+-bound synaptotagmin-1 SNARE complex (long unit cell form) - from synchrotron diffraction | | Descriptor: | CALCIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Lyubimov, A.Y, Uervirojnangkoorn, M, Zhou, Q, Zhao, M, Sauter, N.K, Brewster, A.S, Weis, W.I, Brunger, A.T. | | Deposit date: | 2016-06-17 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Advances in X-ray free electron laser (XFEL) diffraction data processing applied to the crystal structure of the synaptotagmin-1 / SNARE complex.
Elife, 5, 2016
|
|
6LID
 
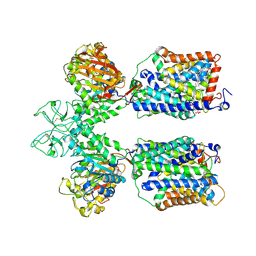 | | Heteromeric amino acid transporter b0,+AT-rBAT complex | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, R.H, Li, Y.N, Lei, J.L, Zhou, Q. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of the human heteromeric amino acid transporter b0,+AT-rBAT.
Sci Adv, 6, 2020
|
|
6LI9
 
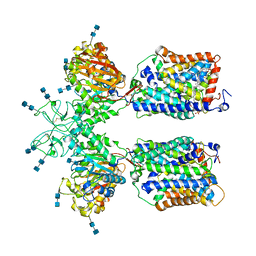 | | Heteromeric amino acid transporter b0,+AT-rBAT complex bound with Arginine | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, ... | | Authors: | Yan, R.H, Li, Y.N, Lei, J.L, Zhou, Q. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structure of the human heteromeric amino acid transporter b0,+AT-rBAT.
Sci Adv, 6, 2020
|
|
1F0C
 
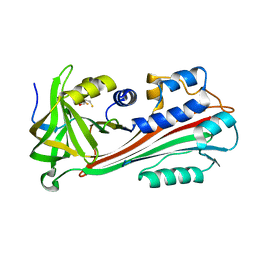 | | STRUCTURE OF THE VIRAL SERPIN CRMA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ICE INHIBITOR | | Authors: | Renatus, M, Zhou, Q, Stennicke, H.R, Snipas, S.J, Turk, D, Bankston, L.A, Liddington, R.C, Salvesen, G.S. | | Deposit date: | 2000-05-15 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the apoptotic suppressor CrmA in its cleaved form.
Structure Fold.Des., 8, 2000
|
|
8QDB
 
 | | Wdyg1p Ser256DHA (PSF) | | Descriptor: | 1,2-ETHANEDIOL, Yellowish-green 1-like protein | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8QD8
 
 | | Wdyg1p in complex with 1,3-Dihydroxynaphthalene | | Descriptor: | 1,2-ETHANEDIOL, Yellowish-green 1-like protein, naphthalene-1,3-diol | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8QD2
 
 | | Ayg1p in complex with 1,3-Dihydroxynaphthalene | | Descriptor: | Pigment biosynthesis protein yellowish-green 1, naphthalene-1,3-diol | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8QD6
 
 | | AntI Ser245DHA (PMSF) | | Descriptor: | ACETATE ION, Photorhabdus luminescens subsp. laumondii TTO1 complete genome segment 15/17, SODIUM ION, ... | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8QD1
 
 | | Ayg1p from A. fumigatus catalyzes polyketide shortening in the biosynthesis of DHN-melanin | | Descriptor: | Pigment biosynthesis protein yellowish-green 1 | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8QD4
 
 | | Ayg1p active site converted to tetrahedral sulfonate ester | | Descriptor: | Pigment biosynthesis protein yellowish-green 1, phenylmethanesulfonic acid | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
