8GVC
 
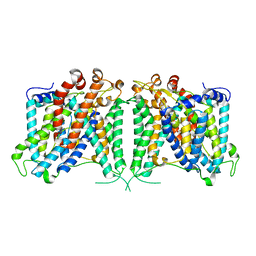 | | The cryo-EM structure of hAE2 with bicarbonate | | Descriptor: | Anion exchange protein 2, BICARBONATE ION | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVF
 
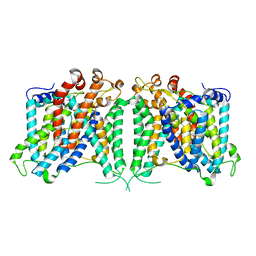 | | The outward-facing structure of hAE2 in basic pH | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVH
 
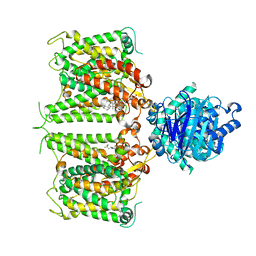 | | Human AE2 in acidic KNO3 | | Descriptor: | Anion exchange protein 2, CHOLESTEROL HEMISUCCINATE | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GV9
 
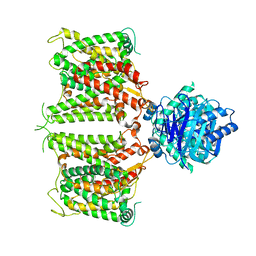 | | The cryo-EM structure of hAE2 with chloride ion | | Descriptor: | Anion exchange protein 2, CHLORIDE ION | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVA
 
 | | The intermediate structure of hAE2 in basic pH | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVE
 
 | | The asymmetry structure of hAE2 | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GV8
 
 | | The cryo-EM structure of hAE2 with DIDS | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
7CY2
 
 | |
7CYR
 
 | |
7CZ2
 
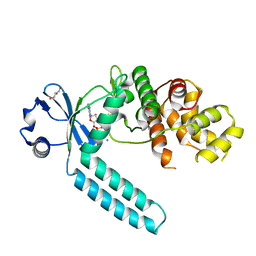 | |
7WAA
 
 | | Crystal structure of MCR-1-S treated by AgNO3 | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, SILVER ION | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2021-12-13 | | Release date: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Re-sensitization of mcr carrying multidrug resistant bacteria to colistin by silver.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7F3X
 
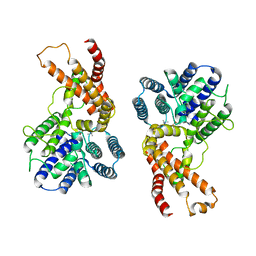 | | Lysophospholipid acyltransferase LPCAT3 in complex with lysophosphatidylcholine | | Descriptor: | LPCAT3, [2-((1-OXODODECANOXY-(2-HYDROXY-3-PROPANYL))-PHOSPHONATE-OXY)-ETHYL]-TRIMETHYLAMMONIUM | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
7EWT
 
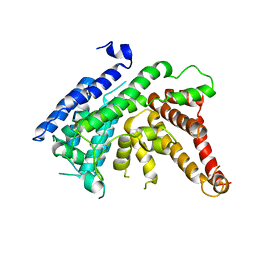 | | The crystal structure of Lysophospholipid acyltransferase LPCAT3 (MOBAT5) in its monomeric and apo form | | Descriptor: | Lysophospholipid acyltransferase 5 | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
7F40
 
 | | Lysophospholipid acyltransferase LPCAT3 in a complex with Arachidonoyl-CoA | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, LPCAT3, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenethioate | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
4Z30
 
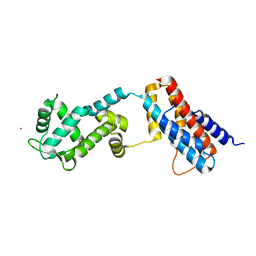 | | Crystal structure of the ROQ domain of human Roquin-2 | | Descriptor: | Roquin-2, UNKNOWN ATOM OR ION | | Authors: | DONG, A, ZHANG, Q, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-30 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | New Insights into the RNA-Binding and E3 Ubiquitin Ligase Activities of Roquins.
Sci Rep, 5, 2015
|
|
3V3Q
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with Ethyl 2-[2,3,4 trimethoxy-6(1-octanoyl)phenyl]acetate | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, SODIUM ION, ... | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
3V3E
 
 | | Crystal Structure of the Human Nur77 Ligand-binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
6LLN
 
 | |
4JGV
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with THPN | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Li, F, Li, A, Tian, X, Wan, W, Wan, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-03-04 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
6LI4
 
 | | Crystal structure of MCR-1-S | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
6LI5
 
 | | Crystal structure of apo-MCR-1-S | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1 | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
1SH4
 
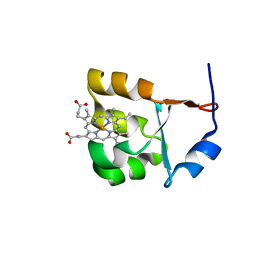 | |
4YWQ
 
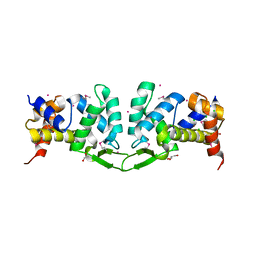 | | Crystal structure of the ROQ domain of human Roquin-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dong, A, Zhang, Q, Li, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-20 | | Release date: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the ROQ domain of human Roquin-1
To be Published
|
|
4Z31
 
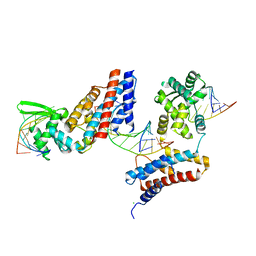 | | Crystal structure of the RC3H2 ROQ domain in complex with stem-loop and double-stranded forms of RNA | | Descriptor: | CHLORIDE ION, RNA (5'-R(*A)-D(P*UP*GP*UP*UP*CP*UP*GP*UP*GP*AP*AP*CP*AP*C)-3'), Roquin-2, ... | | Authors: | DONG, A, ZHANG, Q, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-30 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New Insights into the RNA-Binding and E3 Ubiquitin Ligase Activities of Roquins.
Sci Rep, 5, 2015
|
|
5JJX
 
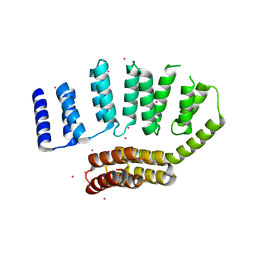 | | Crystal structure of the HAT domain of sart3 | | Descriptor: | CHLORIDE ION, Squamous cell carcinoma antigen recognized by T-cells 3, UNKNOWN ATOM OR ION | | Authors: | DONG, A, ZHANG, Q, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the HAT domain of sart3
to be published
|
|
