8T4Y
 
 | | Human HCN1 F186C S264C C309A bound to cAMP, reconstituted in LMNG + SPL | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Burtscher, V, Mount, J, Cowgill, J, Chang, Y, Bickel, K, Yuan, P, Chanda, B. | | Deposit date: | 2023-06-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for hyperpolarization-dependent opening of human HCN1 channel.
Nat Commun, 15, 2024
|
|
8T50
 
 | | Open human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Burtscher, V, Mount, J, Cowgill, J, Chang, Y, Bickel, K, Yuan, P, Chanda, B. | | Deposit date: | 2023-06-12 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for hyperpolarization-dependent opening of human HCN1 channel.
Nat Commun, 15, 2024
|
|
7UX1
 
 | | EcMscK in an Open Conformation | | Descriptor: | Mechanosensitive channel MscK | | Authors: | Mount, J.W, Yuan, P. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural basis for mechanotransduction in a potassium-dependent mechanosensitive ion channel.
Nat Commun, 13, 2022
|
|
7UW5
 
 | | EcMscK G924S mutant in a closed conformation | | Descriptor: | Mechanosensitive channel MscK | | Authors: | Mount, J.W, Yuan, P. | | Deposit date: | 2022-05-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural basis for mechanotransduction in a potassium-dependent mechanosensitive ion channel.
Nat Commun, 13, 2022
|
|
6M97
 
 | | Crystal structure of the high-affinity copper transporter Ctr1 | | Descriptor: | Chimera protein of High affinity copper uptake protein 1 and Soluble cytochrome b562, HEXATANTALUM DODECABROMIDE, ZINC ION | | Authors: | Ren, F, Yuan, P. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | X-ray structures of the high-affinity copper transporter Ctr1.
Nat Commun, 10, 2019
|
|
6M98
 
 | |
6BBJ
 
 | | Xenopus Tropicalis TRPV4 | | Descriptor: | Transient receptor potential cation channel, subfamily V, member 4 | | Authors: | Deng, Z, Hite, R.K, Yuan, P. | | Deposit date: | 2017-10-18 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6C8G
 
 | | Crystal structure of Transient Receptor Potential (TRP) channel TRPV4 in the presence of barium | | Descriptor: | BARIUM ION, Transient receptor potential cation channel, subfamily V, ... | | Authors: | Deng, Z, Paknejad, N, Maksaev, G, Sala-Rabanal, M, Nichols, C.G, Hite, R.K, Yuan, P. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6.31 Å) | | Cite: | Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6C8H
 
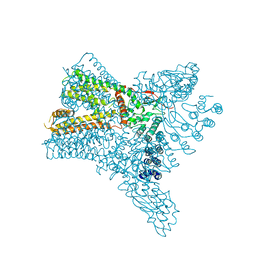 | | Crystal structure of Transient Receptor Potential (TRP) channel TRPV4 in the presence of gadolinium | | Descriptor: | GADOLINIUM ATOM, Transient receptor potential cation channel, subfamily V, ... | | Authors: | Deng, Z, Paknejad, N, Maksaev, G, Sala-Rabanal, M, Nichols, C.G, Hite, R.K, Yuan, P. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6CD2
 
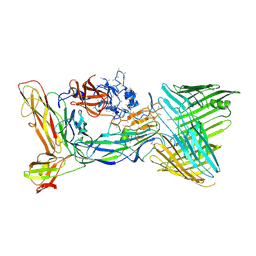 | | Crystal structure of the PapC usher bound to the chaperone-adhesin PapD-PapG | | Descriptor: | Chaperone protein PapD, Outer membrane usher protein PapC, PapGII adhesin protein | | Authors: | Omattage, N.S, Deng, Z, Yuan, P, Hultgren, S.J. | | Deposit date: | 2018-02-07 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for usher activation and intramolecular subunit transfer in P pilus biogenesis in Escherichia coli.
Nat Microbiol, 3, 2018
|
|
6C8F
 
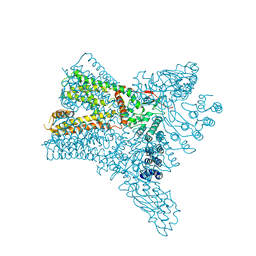 | | Crystal structure of Transient Receptor Potential (TRP) channel TRPV4 in the presence of cesium | | Descriptor: | CESIUM ION, Transient receptor potential cation channel, subfamily V, ... | | Authors: | Deng, Z, Paknejad, N, Maksaev, G, Sala-Rabanal, M, Nichols, C.G, Hite, R.K, Yuan, P. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5IT3
 
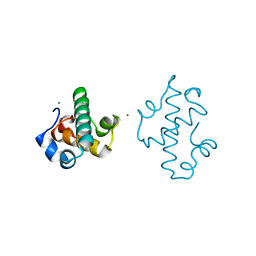 | | Swirm domain of human Lsd1 | | Descriptor: | Lysine-specific histone demethylase 1A, MAGNESIUM ION | | Authors: | Jeffrey, P.D, Yuan, P. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Tlx-interacting peptide of Lsd1 inhibits the proliferation of brain tumor stem cells
To Be Published
|
|
5KUK
 
 | | Crystal Structure of Inward Rectifier Kir2.2 K62W Mutant | | Descriptor: | ATP-sensitive inward rectifier potassium channel 12, DECYL-BETA-D-MALTOPYRANOSIDE, POTASSIUM ION | | Authors: | Lee, S.-J, Ren, F, Heyman, S, Yuan, P, Nichols, C.G. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of control of inward rectifier Kir2 channel gating by bulk anionic phospholipids.
J.Gen.Physiol., 148, 2016
|
|
5KUM
 
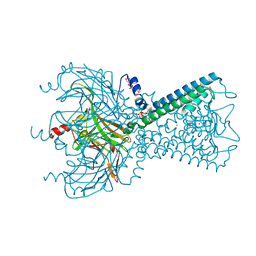 | | Crystal Structure of Inward Rectifier Kir2.2 K62W Mutant In Complex with PIP2 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 12, DECYL-BETA-D-MALTOPYRANOSIDE, POTASSIUM ION, ... | | Authors: | Lee, S.-J, Ren, F, Heyman, S, Yuan, P, Nichols, C.G. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of control of inward rectifier Kir2 channel gating by bulk anionic phospholipids.
J.Gen.Physiol., 148, 2016
|
|
6UW6
 
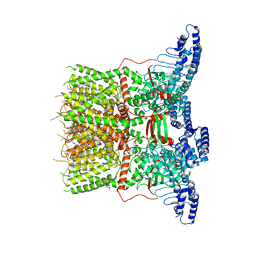 | | Cryo-EM structure of the human TRPV3 K169A mutant determined in lipid nanodisc | | Descriptor: | Transient receptor potential cation channel subfamily V member 3, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, Z, Yuan, P. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Gating of human TRPV3 in a lipid bilayer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UW9
 
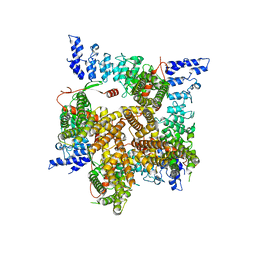 | |
6UW4
 
 | | Cryo-EM structure of human TRPV3 determined in lipid nanodisc | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily V member 3, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, Z, Yuan, P. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Gating of human TRPV3 in a lipid bilayer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6V6D
 
 | | Cryo-EM structure of human pannexin 1 | | Descriptor: | Pannexin-1 | | Authors: | Deng, Z, He, Z, Yuan, P. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-01 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Cryo-EM structures of the ATP release channel pannexin 1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UW8
 
 | |
6UZY
 
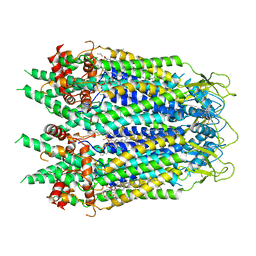 | | Cryo-EM structure of Xenopus tropicalis pannexin 1 | | Descriptor: | HEXADECANE, Pannexin, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, Z, He, Z, Yuan, P. | | Deposit date: | 2019-11-15 | | Release date: | 2020-04-01 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of the ATP release channel pannexin 1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6V5A
 
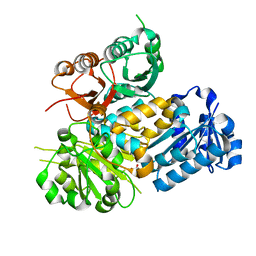 | | Crystal structure of the human BK channel gating ring L390P mutant | | Descriptor: | CALCIUM ION, Calcium-activated potassium channel subunit alpha-1, SULFATE ION | | Authors: | Deng, Z, Yuan, P. | | Deposit date: | 2019-12-03 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coupling of Ca2+and voltage activation in BK channels through the alpha B helix/voltage sensor interface.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VXN
 
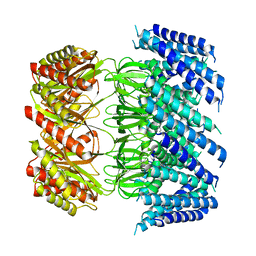 | | Cryo-EM structure of Arabidopsis thaliana MSL1 A320V | | Descriptor: | DODECANE, Mechanosensitive ion channel protein 1, mitochondrial | | Authors: | Deng, Z, Zhang, J, Yuan, P. | | Deposit date: | 2020-02-22 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural mechanism for gating of a eukaryotic mechanosensitive channel of small conductance.
Nat Commun, 11, 2020
|
|
6VXM
 
 | | Cryo-EM structure of Arabidopsis thaliana MSL1 | | Descriptor: | EICOSANE, Mechanosensitive ion channel protein 1, mitochondrial | | Authors: | Deng, Z, Zhang, J, Yuan, P. | | Deposit date: | 2020-02-22 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural mechanism for gating of a eukaryotic mechanosensitive channel of small conductance.
Nat Commun, 11, 2020
|
|
6VXP
 
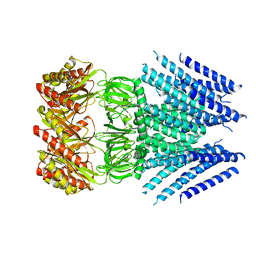 | |
8F7C
 
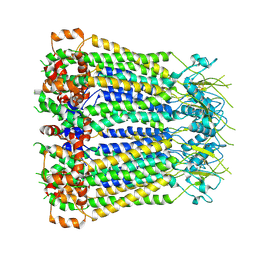 | | Cryo-EM structure of human pannexin 2 | | Descriptor: | Pannexin-2, Soluble cytochrome b562 fusion | | Authors: | He, Z, Yuan, P. | | Deposit date: | 2022-11-18 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural and functional analysis of human pannexin 2 channel.
Nat Commun, 14, 2023
|
|
