7W6V
 
 | | Crystal structure of a dicobalt-substituted small laccase at 2.47 angstrom | | Descriptor: | COBALT (II) ION, Putative copper oxidase | | Authors: | Yang, X, Wu, F, Wu, W, Chen, X, Fan, S, Yu, P, Mao, L. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | A versatile artificial metalloenzyme scaffold enabling direct bioelectrocatalysis in solution.
Sci Adv, 8, 2022
|
|
6VWT
 
 | |
5SWD
 
 | | Structure of the adenine riboswitch aptamer domain in an intermediate-bound state | | Descriptor: | ADENINE, MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, ... | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
7VQ1
 
 | | Structure of Apo-hsTRPM2 channel | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
7VQ2
 
 | | Structure of Apo-hsTRPM2 channel TM domain | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
5SWE
 
 | | Ligand-bound structure of adenine riboswitch aptamer domain converted in crystal from its ligand-free state using ligand mixing serial femtosecond crystallography | | Descriptor: | ADENINE, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
5E54
 
 | | Two apo structures of the adenine riboswitch aptamer domain determined using an X-ray free electron laser | | Descriptor: | MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2015-10-07 | | Release date: | 2016-11-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
4ALW
 
 | | Benzofuropyrimidinone Inhibitors of Pim-1 | | Descriptor: | 8-BROMANYL-2-[(4-METHYLPIPERAZIN-1-YL)METHYL]-3H-[1]BENZOFURO[3,2-D]PYRIMIDIN-4-ONE, IMIDAZOLE, PIM-1 KINASE | | Authors: | Stout, T.J, Adams, L. | | Deposit date: | 2012-03-05 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The Design, Synthesis, and Biological Evaluation of Pim Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4BBE
 
 | | Aminoalkylpyrimidine Inhibitor Complexes with JAK2 | | Descriptor: | N-[4-[2-[(4-morpholin-4-ylphenyl)amino]pyrimidin-4-yl]phenyl]ethanamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Li, J. | | Deposit date: | 2012-09-21 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar and in Vivo Evaluation of 4-Aryl-2-Aminoalkylpyrimidines as Potent and Selective Janus Kinase 2 (Jak2) Inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4ALU
 
 | | Benzofuropyrimidinone Inhibitors of Pim-1 | | Descriptor: | 8-bromo-2-(2-chlorophenyl)[1]benzofuro[3,2-d]pyrimidin-4(3H)-one, IMIDAZOLE, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Stout, T.J, Adams, L. | | Deposit date: | 2012-03-05 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Design, Synthesis, and Biological Evaluation of Pim Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4BBF
 
 | | Aminoalkylpyrimidine Inhibitor Complexes with JAK2 | | Descriptor: | (2R)-N-[4-[2-[(4-morpholin-4-ylphenyl)amino]pyrimidin-4-yl]phenyl]pyrrolidine-2-carboxamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Li, J. | | Deposit date: | 2012-09-21 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sar and in Vivo Evaluation of 4-Aryl-2-Aminoalkylpyrimidines as Potent and Selective Janus Kinase 2 (Jak2) Inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4ALV
 
 | | Benzofuropyrimidinone Inhibitors of Pim-1 | | Descriptor: | 8-bromo-2-{2-chloro-4-[(piperidin-4-ylmethyl)amino]phenyl}[1]benzofuro[3,2-d]pyrimidin-4(3H)-one, IMIDAZOLE, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Stout, T.J, Adams, L. | | Deposit date: | 2012-03-05 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The Design, Synthesis, and Biological Evaluation of Pim Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4ANM
 
 | | Complex of CK2 with a CDC7 inhibitor | | Descriptor: | 8-BROMANYL-2-[[(3S)-3-OXIDANYLPYRROLIDIN-1-YL]METHYL]-3H-[1]BENZOFURO[3,2-D]PYRIMIDIN-4-ONE, CASEIN KINASE II SUBUNIT ALPHA | | Authors: | Stout, T.J. | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Xl413, a Potent and Selective Cdc7 Inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4AW5
 
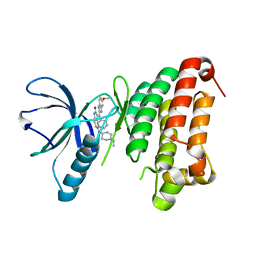 | | Complex of the EphB4 kinase domain with an oxindole inhibitor | | Descriptor: | (3Z)-5-[(1-ethylpiperidin-4-yl)amino]-3-[(5-methoxy-1H-benzimidazol-2-yl)(phenyl)methylidene]-1,3-dihydro-2H-indol-2-one, EPHRIN TYPE-B RECEPTOR 4 | | Authors: | Till, J.H, Stout, T.J. | | Deposit date: | 2012-05-31 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The Design, Synthesis, and Biological Evaluation of Potent Receptor Tyrosine Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6WJR
 
 | |
6WJS
 
 | |
3LQ8
 
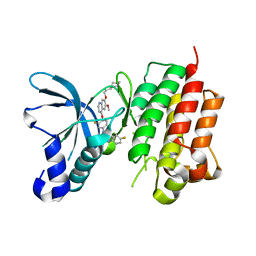 | | Structure of the kinase domain of c-Met bound to XL880 (GSK1363089) | | Descriptor: | Hepatocyte growth factor receptor, N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Lougheed, J.C, Stout, T.J. | | Deposit date: | 2010-02-08 | | Release date: | 2010-05-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Inhibition of tumor cell growth, invasion, and metastasis by EXEL-2880 (XL880, GSK1363089), a novel inhibitor of HGF and VEGF receptor tyrosine kinases.
Cancer Res., 69, 2009
|
|
4XNR
 
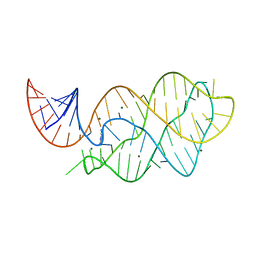 | | Vibrio Vulnificus Adenine Riboswitch Aptamer Domain, Synthesized by Position-selective Labeling of RNA (PLOR), in Complex with Adenine | | Descriptor: | ADENINE, MAGNESIUM ION, Vibrio Vulnificus Adenine Riboswitch | | Authors: | Zhang, J, Liu, Y, Wang, Y.-X, Ferre-D'Amare, A.R. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Synthesis and applications of RNAs with position-selective labelling and mosaic composition.
Nature, 522, 2015
|
|
6VWV
 
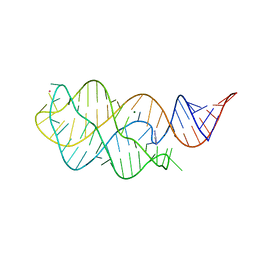 | |
5UZA
 
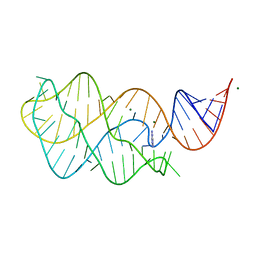 | | Adenine riboswitch aptamer domain labelled with iodo-uridine by position-selective labelling of RNA (PLOR) | | Descriptor: | ADENINE, MAGNESIUM ION, RNA (71-MER) | | Authors: | Liu, Y, Stagno, J.R, Wang, Y.-X. | | Deposit date: | 2017-02-25 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Incorporation of isotopic, fluorescent, and heavy-atom-modified nucleotides into RNAs by position-selective labeling of RNA.
Nat Protoc, 13, 2018
|
|
5YB1
 
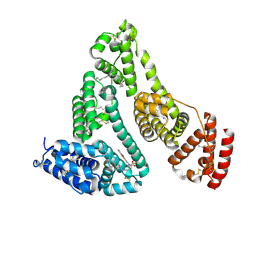 | | Structure and function of human serum albumin-metal agent complex | | Descriptor: | PALMITIC ACID, Serum albumin, ~{N},~{N},12-trimethyl-3$l^{3}-thia-1$l^{4},5,6$l^{4}-triaza-2$l^{3}-cupratricyclo[6.4.0.0^{2,6}]dodeca-1(8),3,6,9,11-pentaen-4-amine | | Authors: | Yang, F, Wang, T, Wang, J, Gou, Y, Zhang, Z.L. | | Deposit date: | 2017-09-02 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Developing an Anticancer Copper(II) Multitarget Pro-Drug Based on the His146 Residue in the IB Subdomain of Modified Human Serum Albumin.
Mol. Pharm., 15, 2018
|
|
8CZJ
 
 | | A bacteria Zrt/Irt-like protein in the apo state | | Descriptor: | Putative membrane protein, SULFATE ION, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Zhang, Y, Hu, J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Structural insights into the elevator-type transport mechanism of a bacterial ZIP metal transporter.
Nat Commun, 14, 2023
|
|
7KD1
 
 | |
8F4O
 
 | | Apo structure of the TPP riboswitch aptamer domain | | Descriptor: | IRIDIUM HEXAMMINE ION, TETRAETHYLENE GLYCOL, TPP riboswitch aptamer domain, ... | | Authors: | Lee, H.-K, Wang, Y.-X, Stagno, J.R. | | Deposit date: | 2022-11-11 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of Escherichia coli thiamine pyrophosphate-sensing riboswitch in the apo state.
Structure, 31, 2023
|
|
2FTU
 
 | | solution structure of domain 3 of RAP | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein, domain 3 | | Authors: | Lee, D, Walsh, J.D, Wang, Y.-X. | | Deposit date: | 2006-01-24 | | Release date: | 2006-05-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | RAP uses a histidine switch to regulate its interaction with LRP in the ER and Golgi.
Mol.Cell, 22, 2006
|
|
