7MJA
 
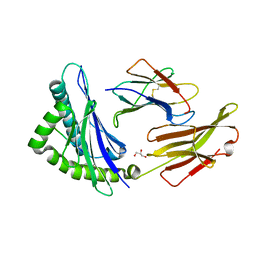 | | HLA-A*24:02 bound to Neuroblastoma derived PHOX2B peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
7MJ8
 
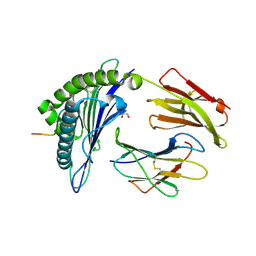 | | HLA-A*02:01 bound to Neuroblastoma Derived IGFBPL1 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Insulin-like growth factor-binding protein-like 1 peptide, ... | | Authors: | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
6MPP
 
 | |
6NPR
 
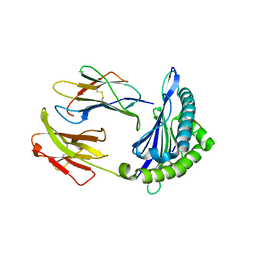 | | Crystal structure of H-2Dd with C84-C139 disulfide in complex with gp120 derived peptide P18-I10 | | Descriptor: | ARG-GLY-PRO-GLY-ARG-ALA-PHE-VAL-THR-ILE, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Toor, J, McShan, A.C, Tripathi, S.M, Sgourakis, N.G. | | Deposit date: | 2019-01-18 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Molecular determinants of chaperone interactions on MHC-I for folding and antigen repertoire selection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6NAN
 
 | | NMR structure determination of Ixolaris and Factor X interaction reveals a noncanonical mechanism of Kunitz inhibition | | Descriptor: | Ixolaris | | Authors: | De Paula, V.S, Sgourakis, N.G, Francischetti, I.M.B, Almeida, F.C.L, Monteiro, R.Q, Valente, A.P. | | Deposit date: | 2018-12-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of Ixolaris and factor X(a) interaction reveals a noncanonical mechanism of Kunitz inhibition.
Blood, 134, 2019
|
|
7RNO
 
 | | Model of the Ac-6-FP/hpMR1/bB2m/TAPBPR complex from integrated docking, NMR and restrained MD | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, N-(6-formyl-4-oxo-3,4-dihydropteridin-2-yl)acetamide, ... | | Authors: | McShan, A.C, Sgourakis, N.G. | | Deposit date: | 2021-07-29 | | Release date: | 2022-05-11 | | Last modified: | 2022-08-10 | | Method: | SOLUTION NMR | | Cite: | TAPBPR employs a ligand-independent docking mechanism to chaperone MR1 molecules.
Nat.Chem.Biol., 18, 2022
|
|
2MJZ
 
 | |
2M89
 
 | | Solution structure of the Aha1 dimer from Colwellia psychrerythraea | | Descriptor: | Aha1 domain protein | | Authors: | Rossi, P, Sgourakis, N.G, Shi, L, Liu, G, Barbieri, C.M, Lee, H, Grant, T.D, Luft, J.R, Xiao, R, Acton, T.B, Montelione, G.T, Snell, E.H, Baker, D, Lange, O.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-09 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | A hybrid NMR/SAXS-based approach for discriminating oligomeric protein interfaces using Rosetta.
Proteins, 83, 2015
|
|
2LPZ
 
 | | Atomic model of the Type-III Secretion System Needle | | Descriptor: | Protein prgI | | Authors: | Loquet, A, Sgourakis, N.G, Gupta, R, Giller, K, Riedel, D, Goosmann, C, Griesinger, C, Kolbe, M.G, Baker, D, Becker, S, Lange, A. | | Deposit date: | 2012-02-21 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic model of the type III secretion system needle.
Nature, 486, 2012
|
|
2MME
 
 | | Hybrid structure of the Shigella flexneri MxiH Type three secretion system needle | | Descriptor: | MxiH | | Authors: | Demers, J.P, Habenstein, B, Loquet, A, Vasa, S.K, Becker, S, Baker, D, Lange, A, Sgourakis, N.G. | | Deposit date: | 2014-03-14 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.7 Å), SOLID-STATE NMR | | Cite: | High-resolution structure of the Shigella type-III secretion needle by solid-state NMR and cryo-electron microscopy.
Nat Commun, 5, 2014
|
|
2MV0
 
 | | Solution NMR Structure of Maltose-binding protein from Escherichia coli, Northeast Structural Genomics Consortium (NESG) Target ER690 | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Rossi, P, Lange, O.F, Sgourakis, N.G, Song, Y, Lee, H, Aramini, J.M, Ertekin, A, Xiao, R, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6B9K
 
 | |
4W4M
 
 | | Crystal structure of PrgK 19-92 | | Descriptor: | Lipoprotein PrgK | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2014-08-15 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Modular Structure of the Inner-Membrane Ring Component PrgK Facilitates Assembly of the Type III Secretion System Basal Body.
Structure, 23, 2015
|
|
4OYC
 
 | |
6E5C
 
 | | Solution NMR structure of a de novo designed double-stranded beta-helix | | Descriptor: | De novo beta protein | | Authors: | Marcos, E, Chidyausiku, T.M, McShan, A, Evangelidis, T, Nerli, S, Sgourakis, N, Tripsianes, K, Baker, D. | | Deposit date: | 2018-07-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | De novo design of a non-local beta-sheet protein with high stability and accuracy.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5IVX
 
 | | Crystal Structure of B4.2.3 T-Cell Receptor and H2-Dd P18-I10 Complex | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Natarajan, K, Jiang, J, Margulies, D. | | Deposit date: | 2016-03-21 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An allosteric site in the T-cell receptor C beta domain plays a critical signalling role.
Nat Commun, 8, 2017
|
|
5IW1
 
 | |
3J6D
 
 | | Model of the PrgH-PrgK periplasmic rings | | Descriptor: | Pathogenicity 1 island effector protein, Protein PrgH | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2014-02-14 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | The Modular Structure of the Inner-Membrane Ring Component PrgK Facilitates Assembly of the Type III Secretion System Basal Body.
Structure, 23, 2015
|
|
4G1I
 
 | |
4G08
 
 | |
4G2S
 
 | |
6O0I
 
 | | NMR ensemble of computationally designed protein XAA | | Descriptor: | Design construct XAA | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6O0C
 
 | | NMR ensemble of computationally designed protein XAA_GVDQ mutant M4L | | Descriptor: | Design construct XAA_GVDQ mutant M4L | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6DMA
 
 | | DHD15_closed | | Descriptor: | DHD15_closed_A, DHD15_closed_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-04 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.363 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DLM
 
 | | DHD127 | | Descriptor: | DHD127_A, DHD127_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-01 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
