2VL6
 
 | | STRUCTURAL ANALYSIS OF THE SULFOLOBUS SOLFATARICUS MCM PROTEIN N- TERMINAL DOMAIN | | Descriptor: | MINICHROMOSOME MAINTENANCE PROTEIN MCM, ZINC ION | | Authors: | Liu, W, Pucci, B, Rossi, M, Pisani, F.M, Ladenstein, R. | | Deposit date: | 2008-01-08 | | Release date: | 2008-04-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of the Sulfolobus Solfataricus Mcm Protein N-Terminal Domain.
Nucleic Acids Res., 36, 2008
|
|
5F8K
 
 | | Crystal structure of the Bac7(1-16) antimicrobial peptide bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Seefeldt, A.C, Graf, M, Perebaskine, N, Nguyen, F, Arenz, S, Mardirossian, M, Scocchi, M, Wilson, D.N, Innis, C.A. | | Deposit date: | 2015-12-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome.
Nucleic Acids Res., 44, 2016
|
|
5FDU
 
 | | Crystal structure of the Metalnikowin I antimicrobial peptide bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Seefeldt, A.C, Graf, M, Perebaskine, N, Nguyen, F, Arenz, S, Mardirossian, M, Scocchi, M, Wilson, D.N, Innis, C.A. | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome.
Nucleic Acids Res., 44, 2016
|
|
5FDV
 
 | | Crystal structure of the Pyrrhocoricin antimicrobial peptide bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Seefeldt, A.C, Graf, M, Perebaskine, N, Nguyen, F, Arenz, S, Mardirossian, M, Scocchi, M, Wilson, D.N, Innis, C.A. | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome.
Nucleic Acids Res., 44, 2016
|
|
7BGC
 
 | | human butyrylcholinesterase in complex with a tacrine-methylanacardate hybrid inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nachon, F, Salerno, A, Bolognesi, M.L. | | Deposit date: | 2021-01-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sustainable Drug Discovery of Multi-Target-Directed Ligands for Alzheimer's Disease.
J.Med.Chem., 64, 2021
|
|
5LLQ
 
 | |
4ZYG
 
 | |
7P4G
 
 | |
4ZYE
 
 | |
4ZYD
 
 | |
4ZYH
 
 | |
5LBH
 
 | | Crystal structure of Helicobacter cinaedi CAIP | | Descriptor: | CAIP, FE (III) ION | | Authors: | Zanotti, G, Valesse, F, Codolo, G, De Bernard, M. | | Deposit date: | 2016-06-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | The Helicobacter cinaedi antigen CAIP participates in atherosclerotic inflammation by promoting the differentiation of macrophages in foam cells.
Sci Rep, 7, 2017
|
|
1GOW
 
 | |
4X5S
 
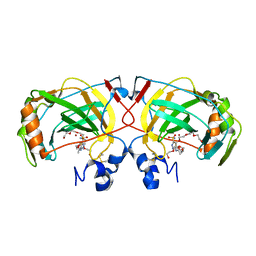 | | The crystal structure of an alpha carbonic anhydrase from the extremophilic bacterium Sulfurihydrogenibium azorense. | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase (Carbonate dehydratase), ... | | Authors: | De Simone, G, Alterio, V, Di Fiore, A. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the most catalytically effective carbonic anhydrase enzyme known, SazCA from the thermophilic bacterium Sulfurihydrogenibium azorense.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8UZD
 
 | |
2CEQ
 
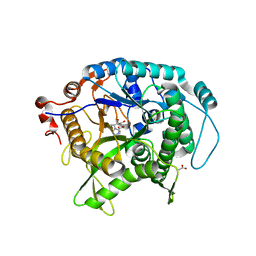 | | Beta-glycosidase from Sulfolobus solfataricus in complex with glucoimidazole | | Descriptor: | ACETATE ION, BETA-GALACTOSIDASE, GLUCOIMIDAZOLE | | Authors: | Gloster, T.M, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2006-02-10 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural, Kinetic, and Thermodynamic Analysis of Glucoimidazole-Derived Glycosidase Inhibitors.
Biochemistry, 45, 2006
|
|
2CER
 
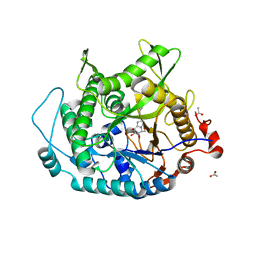 | | Beta-glycosidase from Sulfolobus solfataricus in complex with phenethyl-substituted glucoimidazole | | Descriptor: | (5R,6R,7S,8S)-5-(HYDROXYMETHYL)-2-(2-PHENYLETHYL)-1,5,6,7,8,8A-HEXAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, ACETATE ION, BETA-GLUCOSIDASE A | | Authors: | Gloster, T.M, Roberts, S, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2006-02-10 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural, Kinetic, and Thermodynamic Analysis of Glucoimidazole-Derived Glycosidase Inhibitors.
Biochemistry, 45, 2006
|
|
2CET
 
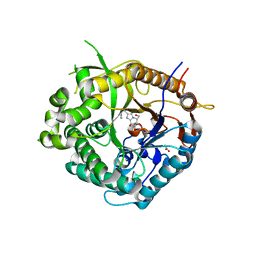 | | Beta-glucosidase from Thermotoga maritima in complex with phenethyl- substituted glucoimidazole | | Descriptor: | (5R,6R,7S,8S)-5-(HYDROXYMETHYL)-2-(2-PHENYLETHYL)-1,5,6,7,8,8A-HEXAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, ACETATE ION, BETA-GLUCOSIDASE A, ... | | Authors: | Gloster, T.M, Roberts, S, Vasella, A, Davies, G.J. | | Deposit date: | 2006-02-10 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural, Kinetic, and Thermodynamic Analysis of Glucoimidazole-Derived Glycosidase Inhibitors.
Biochemistry, 45, 2006
|
|
2CES
 
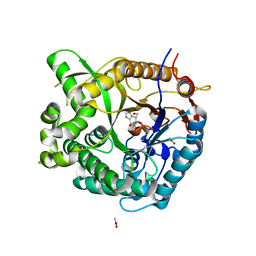 | | Beta-glucosidase from Thermotoga maritima in complex with glucoimidazole | | Descriptor: | ACETATE ION, BETA-GLUCOSIDASE A, CALCIUM ION, ... | | Authors: | Gloster, T.M, Roberts, S, Vasella, A, Davies, G.J. | | Deposit date: | 2006-02-10 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural, Kinetic, and Thermodynamic Analysis of Glucoimidazole-Derived Glycosidase Inhibitors.
Biochemistry, 45, 2006
|
|
1JVB
 
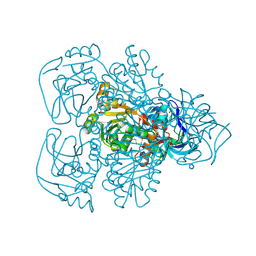 | | ALCOHOL DEHYDROGENASE FROM THE ARCHAEON SULFOLOBUS SOLFATARICUS | | Descriptor: | NAD(H)-DEPENDENT ALCOHOL DEHYDROGENASE, ZINC ION | | Authors: | Esposito, L, Sica, F, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-08-29 | | Release date: | 2002-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the alcohol dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus at 1.85 A resolution.
J.Mol.Biol., 318, 2002
|
|
4G7A
 
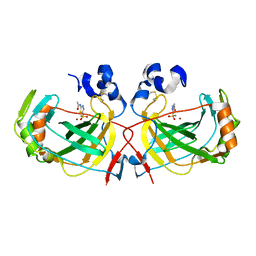 | | The crystal structure of an alpha Carbonic Anhydrase from the extremophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1 | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonate dehydratase, ZINC ION | | Authors: | Di Fiore, A, De Simone, G. | | Deposit date: | 2012-07-20 | | Release date: | 2013-05-29 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of the first `extremo-{alpha}-carbonic anhydrase', a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1NW2
 
 | | The crystal structure of the mutant R82E of Thioredoxin from Alicyclobacillus acidocaldarius | | Descriptor: | ACETATE ION, CACODYLATE ION, THIOREDOXIN, ... | | Authors: | Bartolucci, S, De Simone, G, Galdiero, S, Improta, R, Menchise, V, Pedone, C, Pedone, E, Saviano, M. | | Deposit date: | 2003-02-05 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An integrated structural and computational study of the thermostability of two thioredoxin mutants from Alicyclobacillus acidocaldarius
J.Bacteriol., 185, 2003
|
|
1NSW
 
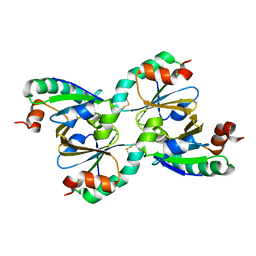 | | The Crystal Structure of the K18G Mutant of the thioredoxin from Alicyclobacillus acidocaldarius | | Descriptor: | THIOREDOXIN | | Authors: | Bartolucci, S, De Simone, G, Galdiero, S, Improta, R, Menchise, V, Pedone, C, Pedone, E, Saviano, M. | | Deposit date: | 2003-01-28 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An integrated structural and computational study of the thermostability of two thioredoxin mutants from Alicyclobacillus acidocaldarius
J.Bacteriol., 185, 2003
|
|
2HM7
 
 | |
1RJW
 
 | |
