3GNY
 
 | | Crystal structure of human alpha-defensin 1 (HNP1) | | Descriptor: | CHLORIDE ION, GLYCEROL, Neutrophil defensin 1 | | Authors: | Pazgier, M, Lu, W.-Y. | | Deposit date: | 2009-03-18 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Through the looking glass, mechanistic insights from enantiomeric human defensins.
J.Biol.Chem., 284, 2009
|
|
4RXZ
 
 | |
2NLF
 
 | |
2NLD
 
 | | Human beta-defensin-1 (Mutant Gln11Ala) | | Descriptor: | ACETATE ION, Beta-defensin 1, SULFATE ION | | Authors: | Lubkowski, J, Pazgier, M. | | Deposit date: | 2006-10-20 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Studies of the Biological Properties of Human beta-Defensin 1.
J.Biol.Chem., 282, 2007
|
|
2NLE
 
 | | Human beta-defensin-1 (Mutant Gln11Ala) | | Descriptor: | ACETATE ION, Beta-defensin 1, SULFATE ION | | Authors: | Lubkowski, J, Pazgier, M. | | Deposit date: | 2006-10-20 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Studies of the Biological Properties of Human beta-Defensin 1.
J.Biol.Chem., 282, 2007
|
|
2NLC
 
 | | Human beta-defensin-1 (mutant Ser8Ala) | | Descriptor: | ACETATE ION, Beta-defensin 1, SULFATE ION | | Authors: | Lubkowski, J, Pazgier, M. | | Deposit date: | 2006-10-20 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Studies of the Biological Properties of Human beta-Defensin 1.
J.Biol.Chem., 282, 2007
|
|
2NLH
 
 | | Human beta-defensin-1 (Mutant GLN24ALA) | | Descriptor: | ACETATE ION, Beta-defensin 1, SULFATE ION | | Authors: | Lubkowski, J, Pazgier, M. | | Deposit date: | 2006-10-20 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Studies of the Biological Properties of Human beta-Defensin 1.
J.Biol.Chem., 282, 2007
|
|
2NLP
 
 | |
2NLS
 
 | |
2NLB
 
 | |
2NLG
 
 | |
2NLQ
 
 | | Human beta-defensin-1 (Mutant Lys31Ala) | | Descriptor: | Beta-defensin 1, GLYCEROL, GLYCINE, ... | | Authors: | Lubkowski, J, Pazgier, M. | | Deposit date: | 2006-10-20 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Studies of the Biological Properties of Human beta-Defensin 1.
J.Biol.Chem., 282, 2007
|
|
7S4S
 
 | |
4FZE
 
 | |
5VK1
 
 | |
5VK0
 
 | |
8TTW
 
 | | Cryo-EM structure of BG505 SOSIP.664 HIV-1 Env trimer in complex with temsavir, 8ANC195, and 10-1074 | | Descriptor: | 1-[4-(benzenecarbonyl)piperazin-1-yl]-2-[4-methoxy-7-(3-methyl-1H-1,2,4-triazol-1-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tolbert, W.D, Pozharski, E, Pazgier, M. | | Deposit date: | 2023-08-15 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
5KJR
 
 | |
8SM2
 
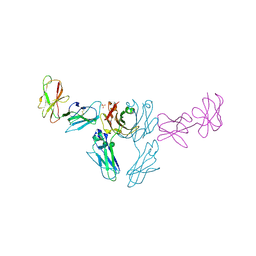 | |
1T3M
 
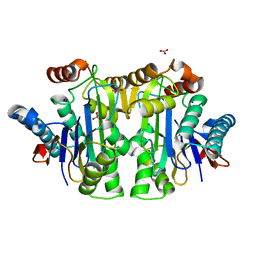 | | Structure of the isoaspartyl peptidase with L-asparaginase activity from E. coli | | Descriptor: | NITRATE ION, Putative L-asparaginase, SODIUM ION | | Authors: | Prahl, A, Pazgier, M, Hejazi, M, Lockau, W, Lubkowski, J. | | Deposit date: | 2004-04-27 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the isoaspartyl peptidase with L-asparaginase activity from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
8GDJ
 
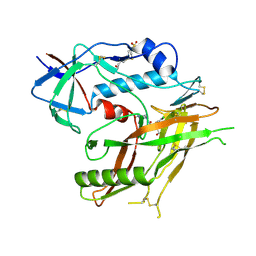 | |
8GDK
 
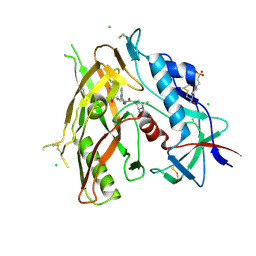 | | Crystal Structure of HIV-1 LM/HT CLADE A/E CRF01 GP120 Core in Complex with TFH-II-151 | | Descriptor: | (3S,5R)-N-(4-chloro-3-fluorophenyl)-5-(hydroxymethyl)-1-[(3R,5S)-3,4,5-trimethylpiperazine-1-carbonyl]piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Nguyen, D.N, Pazgier, M. | | Deposit date: | 2023-03-06 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of HIV-1 LM/HT CLADE A/E CRF01 GP120 Core in Complex with TFH-II-151
To Be Published
|
|
7URQ
 
 | |
7URS
 
 | |
4YC2
 
 | |
