8B4U
 
 | | The crystal structure of PET46, a PETase enzyme from Candidatus bathyarchaeota | | Descriptor: | 1,2-ETHANEDIOL, Alpha/beta hydrolase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Applegate, V, Schumacher, J, Smits, S.H.J. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | An archaeal lid-containing feruloyl esterase degrades polyethylene terephthalate.
Commun Chem, 6, 2023
|
|
1EX9
 
 | | CRYSTAL STRUCTURE OF THE PSEUDOMONAS AERUGINOSA LIPASE COMPLEXED WITH RC-(RP,SP)-1,2-DIOCTYLCARBAMOYL-GLYCERO-3-O-OCTYLPHOSPHONATE | | Descriptor: | CALCIUM ION, LACTONIZING LIPASE, OCTYL-PHOSPHINIC ACID 1,2-BIS-OCTYLCARBAMOYLOXY-ETHYL ESTER | | Authors: | Nardini, M, Lang, D.A, Liebeton, K, Jaeger, K.-E, Dijkstra, B.W. | | Deposit date: | 2000-05-02 | | Release date: | 2000-10-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of pseudomonas aeruginosa lipase in the open conformation. The prototype for family I.1 of bacterial lipases.
J.Biol.Chem., 275, 2000
|
|
6Z68
 
 | | A novel metagenomic alpha/beta-fold esterase | | Descriptor: | Acetyl esterase/lipase, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Bollinger, A, Thies, S, Hoeppner, A, Kobus, S, Jaeger, K.-E, Smits, S.H.J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of a novel family IV esterase in free and substrate-bound form.
Febs J., 288, 2021
|
|
6GSF
 
 | | Solution structure of lipase binding domain LID1 of foldase from Pseudomonas aeruginosa | | Descriptor: | Lipase chaperone | | Authors: | Viegas, A, Jaeger, K.-E, Etzkorn, M, Gohlke, H, Verma, N, Dollinger, P, Kovacic, F. | | Deposit date: | 2018-06-14 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic insights revealing how lipase binding domain MD1 of Pseudomonas aeruginosa foldase affects lipase activation.
Sci Rep, 10, 2020
|
|
6RHF
 
 | | Structure of Chloroflexus aggregans Cagg_3753 LOV domain C85A variant (CagFbFP) | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase | | Authors: | Nazarenko, V.V, Remeeva, A, Yudenko, A, Kovalev, K, Gordeliy, V, Gushchin, I. | | Deposit date: | 2019-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | A thermostable flavin-based fluorescent protein from Chloroflexus aggregans: a framework for ultra-high resolution structural studies.
Photochem. Photobiol. Sci., 18, 2019
|
|
6RHG
 
 | | Structure of Chloroflexus aggregans Cagg_3753 LOV domain | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase | | Authors: | Nazarenko, V.V, Remeeva, A, Yudenko, A, Kovalev, K, Gordeliy, V, Gushchin, I. | | Deposit date: | 2019-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | A thermostable flavin-based fluorescent protein from Chloroflexus aggregans: a framework for ultra-high resolution structural studies.
Photochem. Photobiol. Sci., 18, 2019
|
|
5LUV
 
 | |
5J4E
 
 | |
5J3W
 
 | |
2QUB
 
 | |
2QUA
 
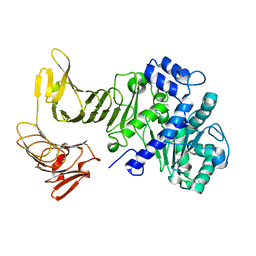 | | Crystal structure of LipA from Serratia marcescens | | Descriptor: | CALCIUM ION, Extracellular lipase | | Authors: | Meier, R, Baumann, U. | | Deposit date: | 2007-08-04 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A calcium-gated lid and a large beta-roll sandwich are revealed by the crystal structure of extracellular lipase from Serratia marcescens.
J.Biol.Chem., 282, 2007
|
|
7R4S
 
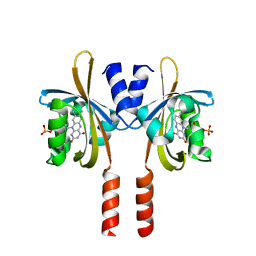 | |
7R56
 
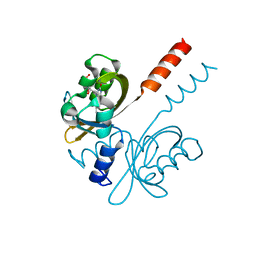 | |
7R5N
 
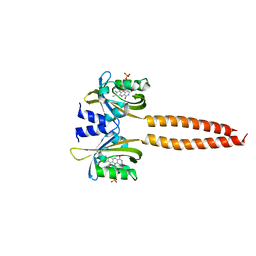 | |
7A6P
 
 | |
6GBV
 
 | |
6GAY
 
 | |
6GB3
 
 | |
6GBA
 
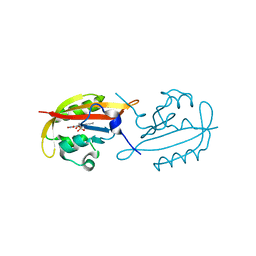 | |
4Q3M
 
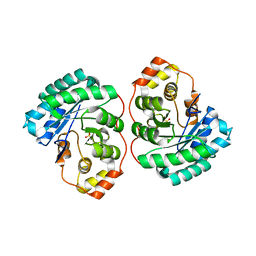 | | Crystal structure of MGS-M4, an aldo-keto reductase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | MGS-M4, SODIUM ION, SULFATE ION | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3K
 
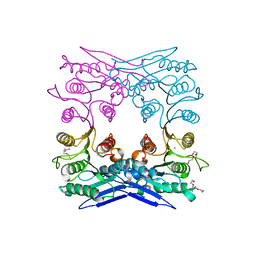 | | Crystal structure of MGS-M1, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | CHLORIDE ION, FLUORIDE ION, MGS-M1, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3L
 
 | | Crystal structure of MGS-M2, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | GLYCEROL, MGS-M2 | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3N
 
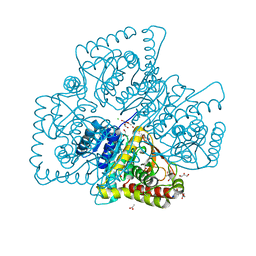 | | Crystal structure of MGS-M5, a lactate dehydrogenase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3O
 
 | | Crystal structure of MGS-MT1, an alpha/beta hydrolase enzyme from a Lake Matapan deep-sea metagenome library | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
