8IEN
 
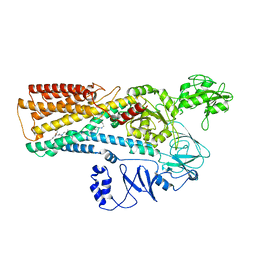 | | Cryo-EM structure of ATP13A2 in the E2-Pi state | | Descriptor: | MAGNESIUM ION, Polyamine-transporting ATPase 13A2, SPERMINE, ... | | Authors: | Liu, Z.M, Mu, J.Q, Xue, C.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Conformational cycle of human polyamine transporter ATP13A2.
Nat Commun, 14, 2023
|
|
8IER
 
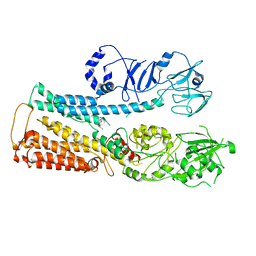 | |
8IES
 
 | | Cryo-EM structure of ATP13A2 in the E1P-ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Polyamine-transporting ATPase 13A2, ... | | Authors: | Liu, Z.M, Mu, J.Q, Xue, C.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Conformational cycle of human polyamine transporter ATP13A2.
Nat Commun, 14, 2023
|
|
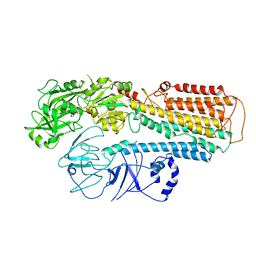
8IEL
 
 | |
8IEO
 
 | | Cryo-EM structure of ATP13A2 in the nominal E1P state | | Descriptor: | MAGNESIUM ION, Polyamine-transporting ATPase 13A2, SPERMINE, ... | | Authors: | Liu, Z.M, Mu, J.Q, Xue, C.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Conformational cycle of human polyamine transporter ATP13A2.
Nat Commun, 14, 2023
|
|
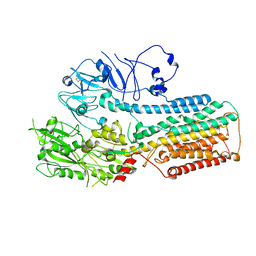
8IEK
 
 | |
4K3J
 
 | | Crystal structure of Onartuzumab Fab in complex with MET and HGF-beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor beta chain, ... | | Authors: | Ma, X, Starovasnik, M.A. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Monovalent antibody design and mechanism of action of onartuzumab, a MET antagonist with anti-tumor activity as a therapeutic agent.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8VEY
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA and TNG908 | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
8VEU
 
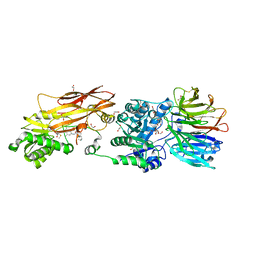 | |
8VET
 
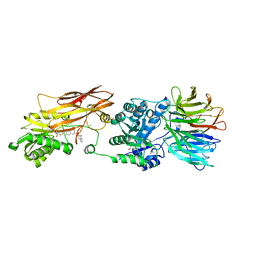 | |
8VEX
 
 | |
8VEW
 
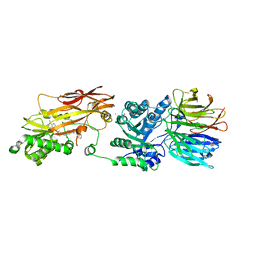 | | Crystal structure of PRMT5:MEP50 in complex with MTA and oxamide compound 24 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5-{2-[(2R,5S)-5-methyl-2-phenylpiperidin-1-yl](oxo)acetamido}pyridine-3-carboxamide, CHLORIDE ION, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
8VEO
 
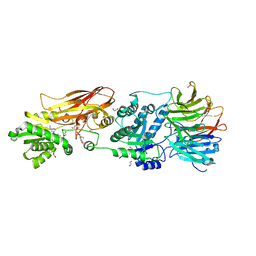 | | Crystal structure of PRMT5:MEP50 in complex with MTA | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
2YB6
 
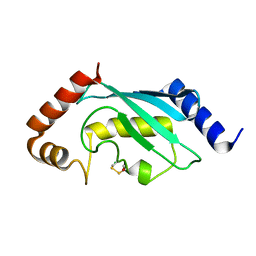 | | Native human Rad6 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, UBIQUITIN-CONJUGATING ENZYME E2 B | | Authors: | Hibbert, R.G, Sixma, T.K. | | Deposit date: | 2011-03-02 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | E3 Ligase Rad18 Promotes Monoubiquitination Rather Than Ubiquitin Chain Formation by E2 Enzyme Rad6.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2YBF
 
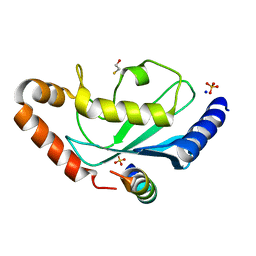 | | Complex of Rad18 (Rad6 binding domain) with Rad6b | | Descriptor: | BETA-MERCAPTOETHANOL, E3 UBIQUITIN-PROTEIN LIGASE RAD18, SODIUM ION, ... | | Authors: | Hibbert, R.G, Sixma, T.K. | | Deposit date: | 2011-03-08 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | E3 Ligase Rad18 Promotes Monoubiquitination Rather Than Ubiquitin Chain Formation by E2 Enzyme Rad6.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
4P59
 
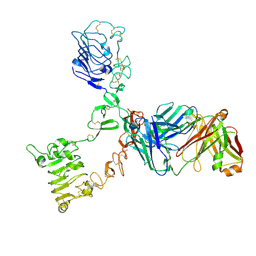 | | HER3 extracellular domain in complex with Fab fragment of MOR09825 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MOR09825 Fab fragment heavy chain, ... | | Authors: | Sprague, E.R. | | Deposit date: | 2014-03-15 | | Release date: | 2014-04-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An antibody that locks HER3 in the inactive conformation inhibits tumor growth driven by HER2 or neuregulin.
Cancer Res., 73, 2013
|
|
3SD5
 
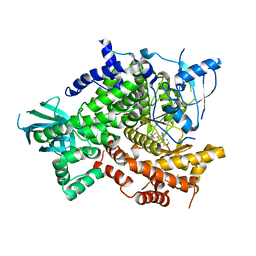 | | Crystal Structure of PI3K gamma with 5-(2,4-dimorpholinopyrimidin-6-yl)-4-(trifluoromethyl)pyridin-2-amine | | Descriptor: | 5-[2,6-di(morpholin-4-yl)pyrimidin-4-yl]-4-(trifluoromethyl)pyridin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2011-06-08 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Identification and Characterization of NVP-BKM120, an Orally Available Pan-Class I PI3-Kinase Inhibitor.
Mol.Cancer Ther., 11, 2012
|
|
3OSV
 
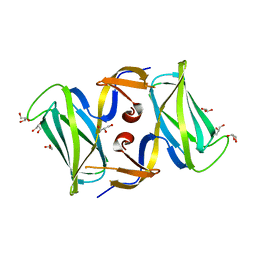 | | The crytsal structure of FLGD from P. Aeruginosa | | Descriptor: | Flagellar basal-body rod modification protein FlgD, GLYCEROL | | Authors: | Wang, D, Luo, M, Niu, S. | | Deposit date: | 2010-09-10 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a novel dimer form of FlgD from P. aeruginosa PAO1
Proteins, 79, 2011
|
|
4FF5
 
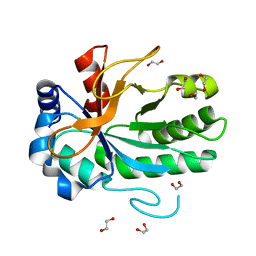 | |
7WS6
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 510A5 heavy chain, 510A5 light chain, Spike protein S1 | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WSA
 
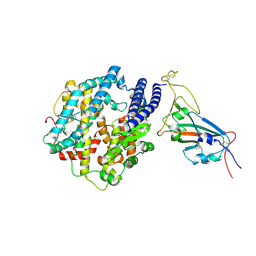 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS2
 
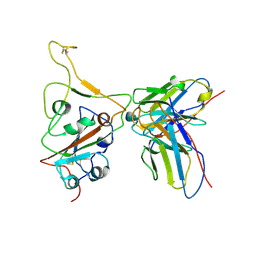 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 510A5 heavy chain, 510A5 light chain, Spike protein S1 | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS8
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
