4LTZ
 
 | | F95M Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.448 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LUU
 
 | | V329A Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LXW
 
 | | L72V Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-30 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LZ3
 
 | | F95H Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, HYDROGENPHOSPHATE ION, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LZ0
 
 | | A236G Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
7SIE
 
 | | Structure of AAP A-domain (residues 351-605) from Staphylococcus epidermidis | | Descriptor: | Accumulation associated protein, CALCIUM ION, CHLORIDE ION | | Authors: | Atkin, K.E, Brentnall, A.S, Dodson, E.J, Whelan, F, Clark, L, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
7SJK
 
 | | Structure of PLS A-domain (residues 391-656) from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Pls Plasmin sensitive surface protein | | Authors: | Clark, L, Whelan, F, Atkin, K.E, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-18 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.208 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
7SMH
 
 | | Structure of SASG A-domain (residues 163-419) from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Surface protein G | | Authors: | Atkin, K.E, Whelan, F, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
1IDK
 
 | | PECTIN LYASE A | | Descriptor: | PECTIN LYASE A | | Authors: | Mayans, O, Scott, M, Connerton, I, Gravesen, T, Benen, J, Visser, J, Pickersgill, R, Jenkins, J. | | Deposit date: | 1996-10-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergence in the substrate-binding clefts of pectin and pectate lyases.
Structure, 5, 1997
|
|
1IDJ
 
 | | PECTIN LYASE A | | Descriptor: | PECTIN LYASE A | | Authors: | Mayans, O, Scott, M, Connerton, I, Gravesen, T, Benen, J, Visser, J, Pickersgill, R, Jenkins, J. | | Deposit date: | 1996-10-04 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergence in the substrate-binding clefts of pectin and pectate lyases.
Structure, 5, 1997
|
|
6R1N
 
 | | Crystal structure of S. aureus seryl-tRNA synthetase complexed to seryl sulfamoyl adenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Salimraj, R, Cain, R, Roper, D.I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Guided Enhancement of Selectivity of Chemical Probe Inhibitors Targeting Bacterial Seryl-tRNA Synthetase.
J.Med.Chem., 62, 2019
|
|
6R1O
 
 | | Crystal structure of E. coli seryl-tRNA synthetase complexed to a seryl sulfamoyl adenosine derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Salimraj, R, Cain, R, Roper, D.I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Enhancement of Selectivity of Chemical Probe Inhibitors Targeting Bacterial Seryl-tRNA Synthetase.
J.Med.Chem., 62, 2019
|
|
6R1M
 
 | | Crystal structure of E. coli seryl-tRNA synthetase complexed to seryl sulfamoyl adenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, PHOSPHATE ION, ... | | Authors: | Salimraj, R, Cain, R, Roper, D.I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Enhancement of Selectivity of Chemical Probe Inhibitors Targeting Bacterial Seryl-tRNA Synthetase.
J.Med.Chem., 62, 2019
|
|
5OYM
 
 | | HIV Integrase Binding Domain of Lens Epithelium-Derived Growth Factor | | Descriptor: | PC4 and SFRS1-interacting protein | | Authors: | Rabbitts, T.H, Phillips, S.E.V, Cruz-Migoni, A, Carr, S.B, Hannon, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cloning, purification and structure determination of the HIV integrase-binding domain of lens epithelium-derived growth factor.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6FB4
 
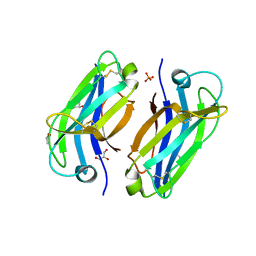 | | human KIBRA C2 domain mutant C771A | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein KIBRA | | Authors: | Crennell, S.J, Posner, M.G, Bagby, S. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.415631 Å) | | Cite: | Distinctive phosphoinositide- and Ca2+-binding properties of normal and cognitive performance-linked variant forms of KIBRA C2 domain.
J. Biol. Chem., 293, 2018
|
|
6FJD
 
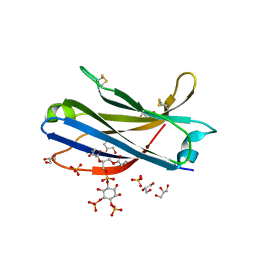 | | Human KIBRA C2 domain mutant C771A in complex with phosphatidylinositol 4,5-bisphosphate | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, GLYCEROL, Protein KIBRA, ... | | Authors: | Crennell, S.J, Posner, M.G, Bagby, S. | | Deposit date: | 2018-01-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Distinctive phosphoinositide- and Ca2+-binding properties of normal and cognitive performance-linked variant forms of KIBRA C2 domain.
J. Biol. Chem., 293, 2018
|
|
6FJC
 
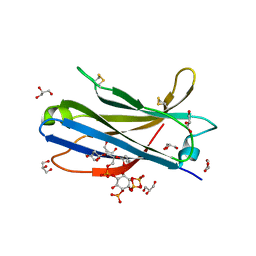 | | Human KIBRA C2 domain mutant C771A in complex with phosphatidylinositol 3,4,5-trisphosphate | | Descriptor: | (2R)-3-{[(S)-{[(2S,3R,5S,6S)-2,6-DIHYDROXY-3,4,5-TRIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-(1-HYDROXY BUTOXY)PROPYL BUTYRATE, GLYCEROL, Protein KIBRA, ... | | Authors: | Crennell, S.J, Posner, M.G, Bagby, S. | | Deposit date: | 2018-01-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Distinctive phosphoinositide- and Ca2+-binding properties of normal and cognitive performance-linked variant forms of KIBRA C2 domain.
J. Biol. Chem., 293, 2018
|
|
6FD0
 
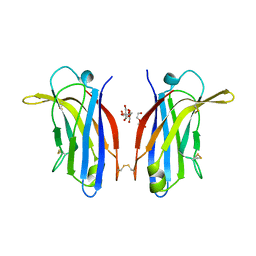 | | Human KIBRA C2 domain mutant M734I S735A | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Protein KIBRA | | Authors: | Crennell, S.J, Posner, M.G, Bagby, S. | | Deposit date: | 2017-12-21 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.64215851 Å) | | Cite: | Distinctive phosphoinositide- and Ca2+-binding properties of normal and cognitive performance-linked variant forms of KIBRA C2 domain.
J. Biol. Chem., 293, 2018
|
|
1WUT
 
 | | Acyl Ureas as Human Liver Glycogen Phosphorylase Inhibitors for the Treatment of Type 2 Diabetes | | Descriptor: | 7-[2,6-DICHLORO-4-({[(2-CHLOROBENZOYL)AMINO]CARBONYL}AMINO)PHENOXY]HEPTANOIC ACID, Glycogen phosphorylase, muscle form, ... | | Authors: | Klabunde, T, Wendt, K.U, Kadereit, D, Brachvogel, V, Burger, H.-J, Herling, A.W, Oikonomakos, N.G, Kosmopoulou, M.N, Schmoll, D, Sarubbi, E, von Roedern, E, Schonafinger, K, Defossa, E. | | Deposit date: | 2004-12-08 | | Release date: | 2005-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystallographic studies on acyl ureas, a new class of glycogen phosphorylase inhibitors, as potential antidiabetic drugs
Protein Sci., 14, 2005
|
|
1GOK
 
 | |
1GOQ
 
 | |
1GOO
 
 | | Thermostable xylanase I from Thermoascus aurantiacus - Cryocooled glycerol complex | | Descriptor: | ENDO-1,4-BETA-XYLANASE, GLYCEROL | | Authors: | Eckert, K, Andrei, C, Larsen, S, Lo Leggio, L. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Substrate Specificity and Subsite Mobility in T. Aurantiacus Xylanase 10A
FEBS Lett., 509, 2001
|
|
1GOM
 
 | |
8C0D
 
 | | UFL1/DDRGK1 bound to UFC1 | | Descriptor: | DDRGK domain-containing protein 1, E3 UFM1-protein ligase 1, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Magnussen, H.M, Kulathu, Y. | | Deposit date: | 2022-12-16 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.564 Å) | | Cite: | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
8BZR
 
 | | UFC1-UFM1 conjugate | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin-fold modifier 1, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Magnussen, H.M, Kulathu, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
