7A5Q
 
 | | Crystal structure of VcSiaP bound to sialic acid | | Descriptor: | DctP family TRAP transporter solute-binding subunit, GLYCEROL, N-acetyl-beta-neuraminic acid, ... | | Authors: | Schneberger, N, Peter, M.F, Hagelueken, G. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Triggering Closure of a Sialic Acid TRAP Transporter Substrate Binding Protein through Binding of Natural or Artificial Substrates.
J.Mol.Biol., 433, 2020
|
|
8CP7
 
 | |
5LTC
 
 | |
7BAH
 
 | | Structure of RIG-I CTD bound to OH-RNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, RNA (5'-R(*GP*AP*CP*GP*CP*UP*AP*GP*CP*GP*UP*C)-3'), ZINC ION | | Authors: | Anand, K, Hagelueken, G, Fusshoeller, D, Geyer, M. | | Deposit date: | 2020-12-15 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A conserved isoleucine in the RNA sensor RIG-I controls immune tolerance to mitochondrial RNA
To Be Published
|
|
7BAI
 
 | |
9FVB
 
 | |
9FVE
 
 | |
9FVC
 
 | |
2X0F
 
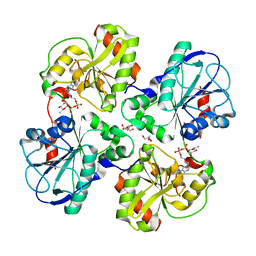 | | Structure of WsaF in complex with dTDP-beta-L-Rha | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, GLYCEROL, WSAF | | Authors: | Steiner, K, Hagelueken, G, Naismith, J.H. | | Deposit date: | 2009-12-08 | | Release date: | 2010-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Substrate Binding in Wsaf, a Rhamnosyltransferase from Geobacillus Stearothermophilus.
J.Mol.Biol., 397, 2010
|
|
7QE5
 
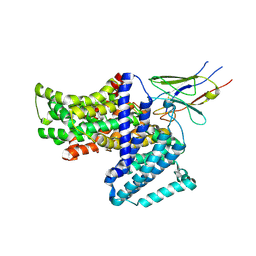 | |
8B0R
 
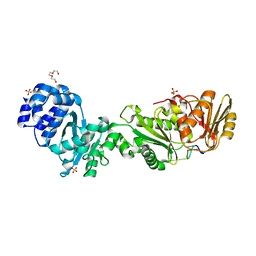 | | Structure of the CalpL/cA4 complex | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), SMODS-associated and fused to various effectors domain-containing protein, SULFATE ION, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
8A90
 
 | | Crystal structure of FrsH | | Descriptor: | ACETATE ION, FE (III) ION, GLYCEROL, ... | | Authors: | Schneberger, N, Wirtz, D.A, Cruesemann, M, Hagelueken, G. | | Deposit date: | 2022-06-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Adenylation Domain-Guided Recruitment of Trans- Acting Nonheme Monooxygenases in Nonribosomal Peptide Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
7QDA
 
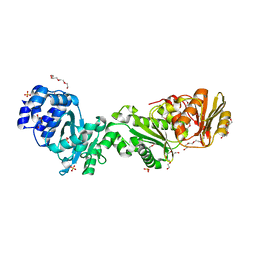 | | Crystal structure of CalpL | | Descriptor: | CalpL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
8B0U
 
 | | Structure of the CalpL/T10 complex | | Descriptor: | CalpT10, GLYCEROL, SAVED domain-containing protein, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
8CMU
 
 | | High resolution structure of the coagulation Factor XIII A2B2 heterotetramer complex. | | Descriptor: | Coagulation factor XIII A chain, Coagulation factor XIII B chain | | Authors: | Singh, S, Urgular, D, Hagelueken, G, Geyer, M, Biswas, A. | | Deposit date: | 2023-02-21 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Cryo-EM structure of the human native plasma coagulation factor XIII complex.
Blood, 2024
|
|
8CMT
 
 | | Structure of the plasma coagulation Factor XIII A2B2 heterotetrameric complex. | | Descriptor: | Coagulation factor XIII A chain, Coagulation factor XIII B chain | | Authors: | Singh, S, Ugurlar, D, Hagelueken, G, Geyer, M, Biswas, A. | | Deposit date: | 2023-02-21 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Cryo-EM structure of the human native plasma coagulation factor XIII complex.
Blood, 2024
|
|
7Z1X
 
 | |
7PZC
 
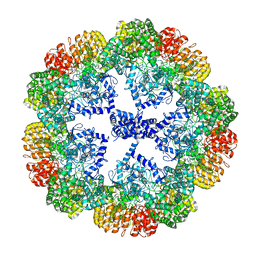 | | Cryo-EM structure of the NLRP3 decamer bound to the inhibitor CRID3 | | Descriptor: | 1-(1,2,3,5,6,7-hexahydro-s-indacen-4-yl)-3-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Hochheiser, I.V, Pilsl, M, Hagelueken, G, Engel, C, Geyer, M. | | Deposit date: | 2021-10-12 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3.
Nature, 604, 2022
|
|
7PZD
 
 | | Cryo-EM structure of the NLRP3 PYD filament | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Hochheiser, I.V, Hagelueken, G, Behrmann, H, Behrmann, E, Geyer, M. | | Deposit date: | 2021-10-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Directionality of PYD filament growth determined by the transition of NLRP3 nucleation seeds to ASC elongation.
Sci Adv, 8, 2022
|
|
2BGC
 
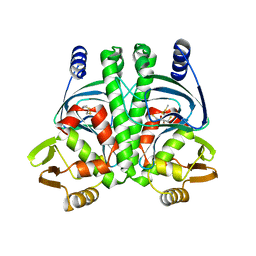 | | PrfA-G145S, a constitutive active mutant of the Transcriptional Regulator In L.monocytogenes | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, PRFA | | Authors: | Eiting, M, Hagelueken, G, Schubert, W.-D, Heinz, D.W. | | Deposit date: | 2004-12-20 | | Release date: | 2005-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Mutation G145S in Prfa, a Key Virulence Regulator of Listeria Monocytogenes, Increases DNA-Binding Affinity by Stabilizing the Hth Motif
Mol.Microbiol., 56, 2005
|
|
2BEO
 
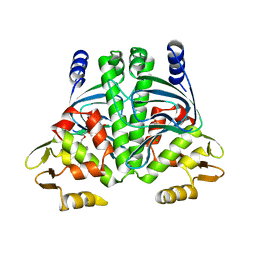 | | PrfA, Transcriptional Regulator In Listeria Monocytogenes | | Descriptor: | ACETAMIDE, CHLORIDE ION, GLUTAMINE, ... | | Authors: | Eiting, M, Hagelueken, G, Schubert, W.-D, Heinz, D.W. | | Deposit date: | 2004-11-26 | | Release date: | 2005-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Mutation G145S in Prfa, a Key Virulence Regulator of Listeria Monocytogenes, Increases DNA-Binding Affinity by Stabilizing the Hth Motif
Mol.Microbiol., 56, 2005
|
|
2X0D
 
 | | APO structure of WsaF | | Descriptor: | GLYCEROL, WSAF | | Authors: | Steiner, K, Hagelueken, G, Naismith, J.H. | | Deposit date: | 2009-12-08 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis of Substrate Binding in Wsaf, a Rhamnosyltransferase from Geobacillus Stearothermophilus.
J.Mol.Biol., 397, 2010
|
|
2X0E
 
 | | Complex structure of WsaF with dTDP | | Descriptor: | GLYCEROL, THYMIDINE-5'-DIPHOSPHATE, WSAF | | Authors: | Steiner, K, Hagelueken, G, Naismith, J.H. | | Deposit date: | 2009-12-08 | | Release date: | 2010-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural Basis of Substrate Binding in Wsaf, a Rhamnosyltransferase from Geobacillus Stearothermophilus.
J.Mol.Biol., 397, 2010
|
|
5OFP
 
 | |
5OFR
 
 | |
