5ISF
 
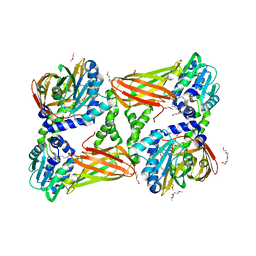 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0705 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0705
To Be Published
|
|
5ISD
 
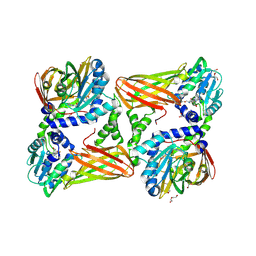 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0592 | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, 5'-S-[(3S)-3-azaniumyl-3-carboxypropyl]-5'-thioadenosine, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0592
To Be Published
|
|
5IS6
 
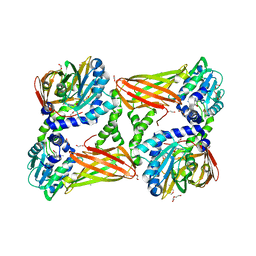 | | Crystal structure of mouse CARM1 in complex with Sinefungin at 2.0 Angstroms resolution | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with Sinefungin at 2.0 Angstroms resolution
To Be Published
|
|
5IS9
 
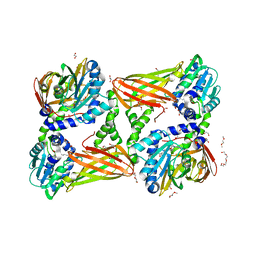 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0375 | | Descriptor: | (2S)-2-amino-4-[(3-{4-[(2S)-2-amino-2-carboxyethyl]-1H-1,2,3-triazol-1-yl}propyl){[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}amino]butanoic acid (non-preferred name), 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0375
To Be Published
|
|
5ISA
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0401 | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azidopropyl)amino]-2-azanyl-butanoic acid, 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0401
To Be Published
|
|
4Z9V
 
 | | TCTP contains a BH3-like domain, which instead of inhibiting, activates Bcl-xL | | Descriptor: | BICARBONATE ION, Bcl-2-like protein 1,APOPTOSIS REGULATOR BCL-XL, SODIUM ION, ... | | Authors: | Cura, V, Agez, M, Thebault, S, Cavarelli, J. | | Deposit date: | 2015-04-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | TCTP contains a BH3-like domain, which instead of inhibiting, activates Bcl-xL.
Sci Rep, 6, 2016
|
|
5LV3
 
 | | Crystal structure of mouse CARM1 in complex with ligand LH1561Br | | Descriptor: | 5-[[2-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethylamino]methyl]-4-azanyl-1-[2-(4-bromanylphenoxy)ethyl]pyrimidin-2-one, Histone-arginine methyltransferase CARM1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
5LV4
 
 | | Crystal structure of mouse PRMT6 in complex with inhibitor LH1236 | | Descriptor: | (2~{R})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[[4-azanyl-1-(methoxymethyl)-2-oxidanylidene-pyrimidin-5-yl]methyl]amino]-2-azanyl-butanoic acid, Protein arginine N-methyltransferase 6 | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
5LV2
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1246 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
5LV5
 
 | | Crystal structure of mouse PRMT6 in complex with inhibitor LH1458 | | Descriptor: | 2-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethyl-[[4-azanyl-1-(methoxymethyl)-2-oxidanylidene-pyrimidin-5-yl]methyl]-[(3~{S})-3-azanyl-4-oxidanyl-4-oxidanylidene-butyl]azanium, Protein arginine N-methyltransferase 6 | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
5TBJ
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1452 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
5TBH
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1236 | | Descriptor: | (2~{R})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[[4-azanyl-1-(methoxymethyl)-2-oxidanylidene-pyrimidin-5-yl]methyl]amino]-2-azanyl-butanoic acid, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
5TBI
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1427 | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-[2-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylamino]ethylamino]-1-(methoxymethyl)pyrimidin-2-one, ... | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
3B3J
 
 | | The 2.55 A crystal structure of the apo catalytic domain of coactivator-associated arginine methyl transferase I(CARM1:28-507, residues 28-146 and 479-507 not ordered) | | Descriptor: | BENZAMIDINE, Histone-arginine methyltransferase CARM1 | | Authors: | Troffer-Charlier, N, Cura, V, Hassenboehler, P, Moras, D, Cavarelli, J. | | Deposit date: | 2007-10-22 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains.
Embo J., 26, 2007
|
|
3B3G
 
 | | The 2.4 A crystal structure of the apo catalytic domain of coactivator-associated arginine methyl transferase I(CARM1,140-480). | | Descriptor: | Histone-arginine methyltransferase CARM1 | | Authors: | Troffer-Charlier, N, Cura, V, Hassenboehler, P, Moras, D, Cavarelli, J. | | Deposit date: | 2007-10-22 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains.
Embo J., 26, 2007
|
|
3B3F
 
 | | The 2.2 A crystal structure of the catalytic domain of coactivator-associated arginine methyl transferase I(CARM1,142-478), in complex with S-adenosyl homocysteine | | Descriptor: | Histone-arginine methyltransferase CARM1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Troffer-Charlier, N, Cura, V, Hassenboehler, P, Moras, D, Cavarelli, J. | | Deposit date: | 2007-10-22 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains.
Embo J., 26, 2007
|
|
4DMA
 
 | | Crystal structure of ERa LBD in complex with RU100132 | | Descriptor: | 2'-bromo-6'-(furan-3-yl)-4'-(hydroxymethyl)biphenyl-4-ol, Estrogen receptor, Nuclear receptor coactivator 1 | | Authors: | Osz, J, Brelivet, Y, Peluso-Iltis, C, Cura, V, Eiler, S, Ruff, M, Bourguet, W, Rochel, N, Moras, D. | | Deposit date: | 2012-02-07 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DM8
 
 | | Crystal structure of RARb LBD in complex with 9cis retinoic acid | | Descriptor: | Nuclear receptor coactivator 1, RETINOIC ACID, Retinoic acid receptor beta | | Authors: | Osz, J, Br livet, Y, Peluso-Iltis, C, Cura, V, Eiler, S, Ruff, M, Bourguet, W, Rochel, N, Moras, D. | | Deposit date: | 2012-02-07 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DM6
 
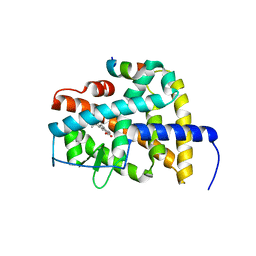 | | Crystal structure of RARb LBD homodimer in complex with TTNPB | | Descriptor: | 4-[(1E)-2-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)PROP-1-ENYL]BENZOIC ACID, Nuclear receptor coactivator 1, Retinoic acid receptor beta | | Authors: | Osz, J, Brelivet, Y, Peluso-Iltis, C, Cura, V, Eiler, S, Ruff, M, Bourguet, W, Rochel, N, Moras, D. | | Deposit date: | 2012-02-07 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GIZ
 
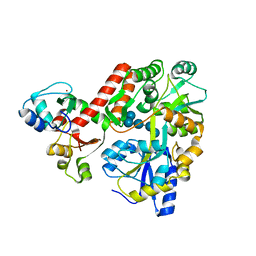 | | Crystal structure of full-length human papillomavirus oncoprotein E6 in complex with LXXLL peptide of ubiquitin ligase E6AP at 2.55 A resolution | | Descriptor: | Maltose-binding periplasmic protein, UBIQUITIN LIGASE EA6P: chimeric protein, Protein E6, ... | | Authors: | McEwen, A.G, Zanier, K, Charbonnier, S, Poussin, P, Cura, V, Vande Pol, S, Trave, G, Cavarelli, J. | | Deposit date: | 2012-08-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins.
Science, 339, 2013
|
|
7QRD
 
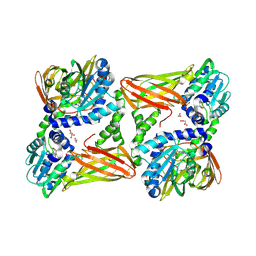 | | Crystal structure of mouse CARM1 in complex with histone H3_10-25 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-01-11 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CARM1 Transition State Mimics
To Be Published
|
|
7QPH
 
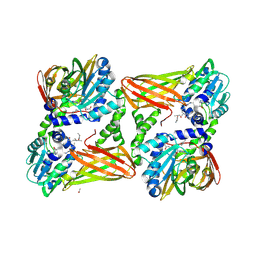 | | Crystal structure of mouse CARM1 in complex with histone H3_22-31 K27 acetylated | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-ETHANEDIOL, Histone H3 22-31 K27 acetylated, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-01-04 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CARM1 Transition State Mimics
To Be Published
|
|
2XPP
 
 | | Crystal structure of a Spt6-Iws1(Spn1) complex from Encephalitozoon cuniculi, Form III | | Descriptor: | CHROMATIN STRUCTURE MODULATOR, IWS1 | | Authors: | Diebold, M.-L, Koch, M, Cura, V, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-27 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
2XPL
 
 | | Crystal structure of Iws1(Spn1) conserved domain from Encephalitozoon cuniculi | | Descriptor: | CHLORIDE ION, IWS1 | | Authors: | Koch, M, Diebold, M.-L, Cura, V, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-27 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
2XPO
 
 | | Crystal structure of a Spt6-Iws1(Spn1) complex from Encephalitozoon cuniculi, Form II | | Descriptor: | CHLORIDE ION, CHROMATIN STRUCTURE MODULATOR, IWS1 | | Authors: | Diebold, M.-L, Koch, M, Cura, V, Moras, D, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-27 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
