2BJW
 
 | | PspF AAA domain | | Descriptor: | PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Beuron, F, Niwa, H, Bordes, P, Wigneshweraraj, S, Keetch, C.A, Robinson, C.V, Buck, M, Zhang, X. | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights Into the Activity of Enhancer-Binding Proteins
Science, 307, 2005
|
|
2BJV
 
 | | Crystal Structure of PspF(1-275) R168A mutant | | Descriptor: | PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Beuron, F, Niwa, H, Bordes, P, Wigneshweraraj, S, Keetch, C.A, Robinson, C.V, Buck, M, Zhang, X. | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights Into the Activity of Enhancer-Binding Proteins
Science, 307, 2005
|
|
3HPN
 
 | | Ligand recognition by A-class EPH receptors: crystal structures of the EPHA2 ligand-binding domain and the EPHA2/EPHRIN-A1 complex | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Himanen, J.P, Goldgur, Y, Miao, H, Myshkin, E, Guo, H, Buck, M, Nguyen, M, Rajashankar, K.R, Wang, B, Nikolov, D.B. | | Deposit date: | 2009-06-04 | | Release date: | 2009-06-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex.
Embo Rep., 10, 2009
|
|
3HEI
 
 | | Ligand Recognition by A-Class Eph Receptors: Crystal Structures of the EphA2 Ligand-Binding Domain and the EphA2/ephrin-A1 Complex | | Descriptor: | Ephrin type-A receptor 2, Ephrin-A1 | | Authors: | Himanen, J.P, Goldgur, Y, Miao, H, Myshkin, E, Guo, H, Buck, M, Nguyen, M, Rajashankar, K.R, Wang, B, Nikolov, D.B. | | Deposit date: | 2009-05-08 | | Release date: | 2009-06-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex.
Embo Rep., 10, 2009
|
|
3H6N
 
 | | Crystal Structure of the ubiquitin-like domain of plexin D1 | | Descriptor: | ARSENIC, Plexin-D1, UNKNOWN ATOM OR ION | | Authors: | Tong, Y, Nedyalkova, L, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Buck, M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-23 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal Structure of the ubiquitin-like domain of plexin D1
To be Published
|
|
3KUZ
 
 | | Crystal structure of the ubiquitin like domain of PLXNC1 | | Descriptor: | Plexin-C1, UNKNOWN ATOM OR ION | | Authors: | Wang, H, Li, B, Tempel, W, Tong, Y, Guan, X, Zhong, N, Crombet, L, MacKenzie, F, Buck, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ubiquitin like domain of PLXNC1
to be published
|
|
2R2O
 
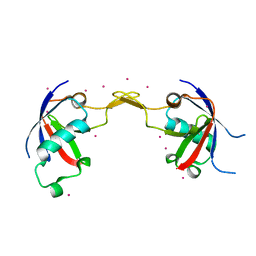 | | Crystal structure of the effector domain of human Plexin B1 | | Descriptor: | Plexin-B1, UNKNOWN ATOM OR ION | | Authors: | Tong, Y, Tempel, W, Shen, L, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-27 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of Rac1, Rnd1, and RhoD to a novel Rho GTPase interaction motif destabilizes dimerization of the plexin-B1 effector domain.
J.Biol.Chem., 282, 2007
|
|
6R9G
 
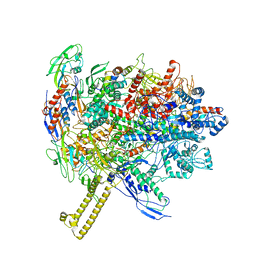 | |
6R9B
 
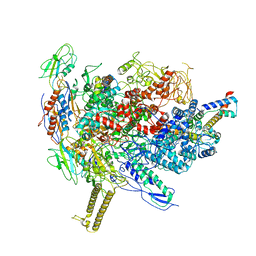 | |
5UI8
 
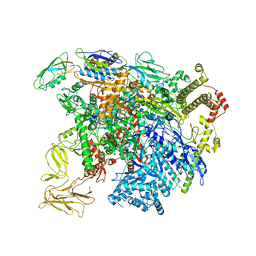 | | structure of sigmaN-holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darst, S.A, Campbell, E.A. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.76 Å) | | Cite: | Crystal structure of Aquifex aeolicus sigma (N) bound to promoter DNA and the structure of sigma (N)-holoenzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WY8
 
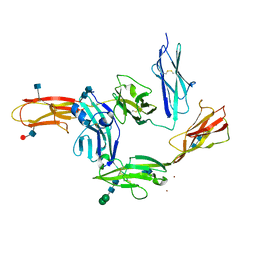 | | Crystal structure of PTP delta Ig1-Ig3 in complex with IL1RAPL1 Ig1-Ig3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein-like 1, ... | | Authors: | Kim, H.M, Won, S.Y, Kim, D. | | Deposit date: | 2017-01-11 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | LAR-RPTP Clustering Is Modulated by Competitive Binding between Synaptic Adhesion Partners and Heparan Sulfate
Front Mol Neurosci, 10, 2017
|
|
9Q91
 
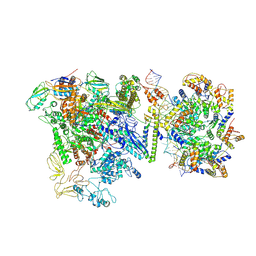 | |
9Q93
 
 | |
9Q94
 
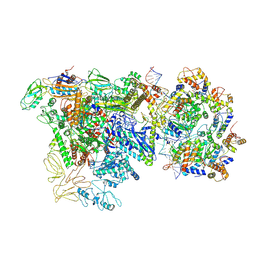 | |
9Q96
 
 | |
9Q97
 
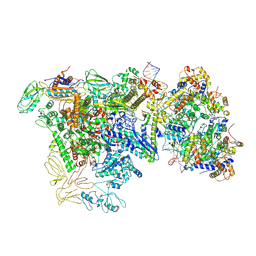 | |
9Q95
 
 | |
9Q98
 
 | |
7QXI
 
 | |
7QWP
 
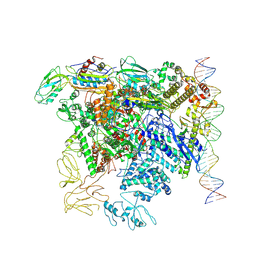 | | CryoEM structure of bacterial transcription close complex (RPc) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Ye, F.Z, Zhang, X.D. | | Deposit date: | 2022-01-25 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanisms of DNA opening revealed in AAA+ transcription complex structures.
Sci Adv, 8, 2022
|
|
7QV9
 
 | |
7BEG
 
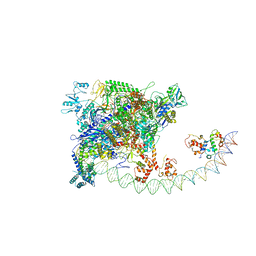 | | Structures of class I bacterial transcription complexes | | Descriptor: | Class I pacrA promoter non-template DNA, Class I pacrA promoter template DNA, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Ye, F.Z, Hao, M, Zhang, X.D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation.
Adv Sci, 9, 2022
|
|
7BEF
 
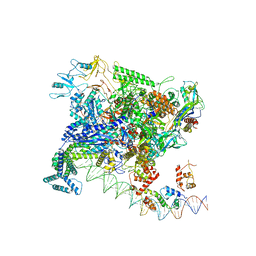 | | Structures of class II bacterial transcription complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Hao, M, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation.
Adv Sci, 9, 2022
|
|
8RE4
 
 | |
8REE
 
 | |
