4OB8
 
 | | Crystal structure of a novel thermostable esterase from Pseudomonas putida ECU1011 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4OU5
 
 | | Crystal structure of esterase rPPE mutant S159A/W187H | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-02-15 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4OU4
 
 | | Crystal structure of esterase rPPE mutant S159A complexed with (S)-Ac-CPA | | Descriptor: | (2S)-(acetyloxy)(2-chlorophenyl)ethanoic acid, Alpha/beta hydrolase fold-3 domain protein | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-02-15 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
5YB5
 
 | | The complex crystal structure of VrEH2 mutant M263N with SNO | | Descriptor: | (S)-PARA-NITROSTYRENE OXIDE, Epoxide hydrolase | | Authors: | Li, F.L, Chen, F.F, Chen, Q, Kong, X.D, Yu, H.L, Xu, J.H. | | Deposit date: | 2017-09-03 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Regioselectivity Engineering of Epoxide Hydrolase: Near-Perfect Enantioconvergence through a Single Site Mutation
Acs Catalysis, 8, 2018
|
|
8WMT
 
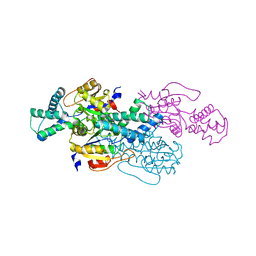 | |
8WNH
 
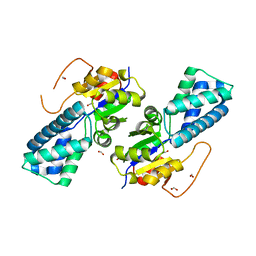 | |
8WWS
 
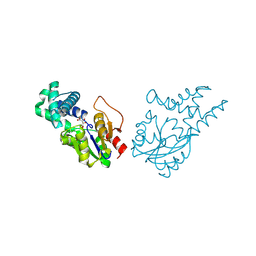 | | Crystal structure of cis-epoxysuccinate hydrolase from Klebsiella oxytoca with L(+)-tartaric acid | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL, CARBONATE ION, ... | | Authors: | Han, Y, Kong, X.D, Li, J, Xu, J.H. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Insights of a cis-Epoxysuccinate Hydrolase Facilitate the Development of Robust Biocatalysts for the Production of l-(+)-Tartrate
Biochemistry, 2024
|
|
4O98
 
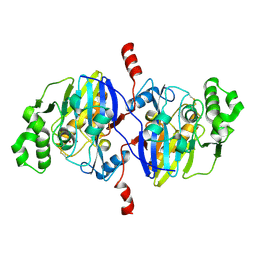 | | Crystal structure of Pseudomonas oleovorans PoOPH mutant H250I/I263W | | Descriptor: | ZINC ION, organophosphorus hydrolase | | Authors: | Luo, X.J, Kong, X.D, Zhao, J, Chen, Q, Zhou, J.H, Xu, J.H. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-03 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Switching a newly discovered lactonase into an efficient and thermostable phosphotriesterase by simple double mutations His250Ile/Ile263Trp
Biotechnol.Bioeng., 111, 2014
|
|
4O08
 
 | |
4NZZ
 
 | |
6LQY
 
 | | Crystal complex of endo-deglycosylated hydroxynitrile lyase isozyme 5 of Prunus communis with benzaldehyde | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, PREDICTED: (R)-mandelonitrile lyase, ... | | Authors: | Zheng, Y.C, Li, F.L, Yu, H.L, Xu, J.H. | | Deposit date: | 2020-01-15 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Structure-Guided Tuning of a Hydroxynitrile Lyase to Accept Rigid Pharmaco Aldehydes.
Acs Catalysis, 2020
|
|
4EEZ
 
 | | Crystal Structure of Lactococcus lactis Alcohol Dehydrogenase variant RE1 | | Descriptor: | Alcohol dehydrogenase 1, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Liu, X, Bastian, S, Snow, C.D, Brustad, E.M, Saleski, T, Xu, J.H, Meinhold, P, Arnold, F.H. | | Deposit date: | 2012-03-28 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided engineering of Lactococcus lactis alcohol dehydrogenase LlAdhA for improved conversion of isobutyraldehyde to isobutanol.
J.Biotechnol., 164, 2012
|
|
4EEX
 
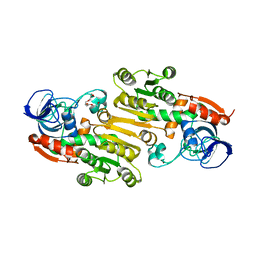 | | Crystal Structure of Lactococcus lactis Alcohol Dehydrogenase | | Descriptor: | Alcohol dehydrogenase 1, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Liu, X, Bastian, S, Snow, C.D, Brustad, E.M, Saleski, T, Xu, J.H, Meinhold, P, Arnold, F.H. | | Deposit date: | 2012-03-28 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided engineering of Lactococcus lactis alcohol dehydrogenase LlAdhA for improved conversion of isobutyraldehyde to isobutanol.
J.Biotechnol., 164, 2012
|
|
5XMD
 
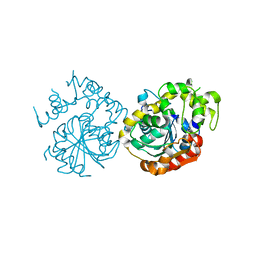 | |
6KBH
 
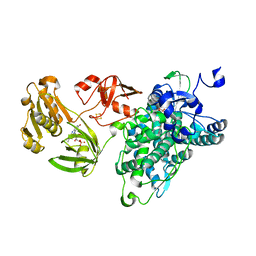 | | Crystal structure of an intact type IV self-sufficient cytochrome P450 monooxygenase | | Descriptor: | Cytochrome P450 monooxygenase, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gong, R, Wu, L.J, Zhang, Y, Liu, Z, Dou, S, Zhang, R.G, Xu, J.H, Tang, C, Zhou, J.H. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an intact type IV self-sufficient cytochrome P450 monooxygenase
To Be Published
|
|
5EIP
 
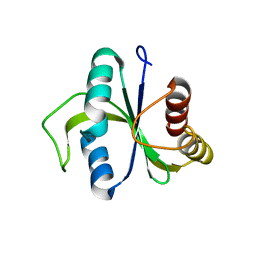 | | apo-structure of YTH domain of SpMmi1 | | Descriptor: | YTH domain-containing protein mmi1 | | Authors: | Wu, B.X, Xu, J.H, Su, S.C, Ma, J.B. | | Deposit date: | 2015-10-30 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insights into the specific recognition of DSR by the YTH domain containing protein Mmi1
Biochem. Biophys. Res. Commun., 491, 2017
|
|
5EIM
 
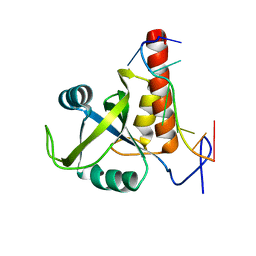 | | YTH domain-containing protein mmi1 and RNA complex | | Descriptor: | RNA (5'-R(*AP*UP*UP*AP*AP*AP*CP*A)-3'), YTH domain-containing protein mmi1 | | Authors: | Wu, B.X, Xu, J.H, Su, S.C, Ma, J.B. | | Deposit date: | 2015-10-30 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the specific recognition of DSR by the YTH domain containing protein Mmi1
Biochem. Biophys. Res. Commun., 491, 2017
|
|
6A37
 
 | |
8J3P
 
 | | Formate dehydrogenase mutant from from Candida dubliniensis M4 complexed with NADP+ | | Descriptor: | Formate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ma, W, Zheng, Y.C, Geng, Q, Chen, C, Xu, J.H. | | Deposit date: | 2023-04-17 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering a Formate Dehydrogenase for NADPH Regeneration.
Chembiochem, 24, 2023
|
|
5GMO
 
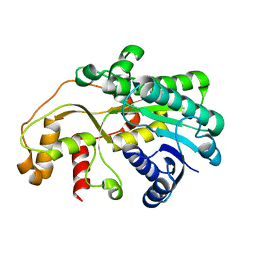 | | X-ray structure of carbonyl reductase SsCR | | Descriptor: | Protein induced by osmotic stress | | Authors: | Shang, Y.P, Yu, H.L, Xu, J.H. | | Deposit date: | 2016-07-14 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Efficient Synthesis of (R)-2-Chloro-1-(2,4-dichlorophenyl)ethanol with a Ketoreductase from Scheffersomyces stipitis CBS 6045
Adv.Synth.Catal., 359, 2017
|
|
6JBY
 
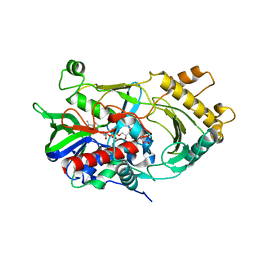 | | Crystal structure of endo-deglycosylated hydroxynitrile lyase isozyme 5 of Prunus communis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zheng, Y.C, Li, F.L, Yu, H.L, Xu, J.H. | | Deposit date: | 2019-01-27 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Tuning of a Hydroxynitrile Lyase to Accept Rigid Pharmaco Aldehydes.
Acs Catalysis, 2020
|
|
5ZFM
 
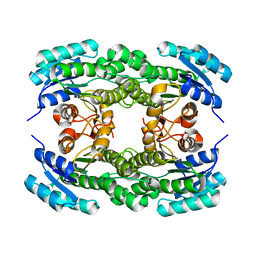 | | Ketoreductase LbCR mutant - M6 | | Descriptor: | 3-oxoacyl-acyl carrier protein reductase | | Authors: | Gong, X.M, Zheng, G.W, Xu, J.H. | | Deposit date: | 2018-03-06 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of an Engineered Ketoreductase with Simultaneously Improved Thermostability and Activity for Making a Bulky Atorvastatin Precursor
Acs Catalysis, 9, 2019
|
|
5ZI0
 
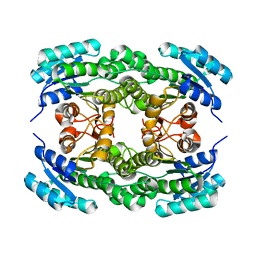 | | Ketoreductase LbCR mutant - M8 | | Descriptor: | 3-oxoacyl-acyl carrier protein reductase | | Authors: | Gong, X.M, Zheng, G.W, Xu, J.H. | | Deposit date: | 2018-03-14 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Development of an Engineered Ketoreductase with Simultaneously Improved Thermostability and Activity for Making a Bulky Atorvastatin Precursor
Acs Catalysis, 9, 2019
|
|
5YWL
 
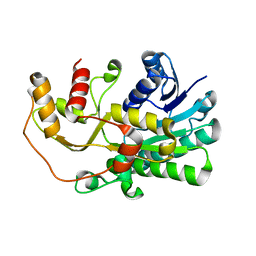 | | SsCR_L211H | | Descriptor: | Protein induced by osmotic stress | | Authors: | Shang, Y.P, Chen, Q, Li, A.T, Yu, H.L, Xu, J.H. | | Deposit date: | 2017-11-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Attenuated substrate inhibition of a haloketone reductase via structure-guided loop engineering.
J.Biotechnol., 308, 2020
|
|
5YWN
 
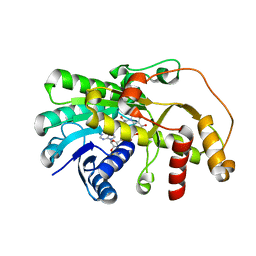 | | SsCR_L211H-NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Protein induced by osmotic stress | | Authors: | Shang, Y.P, Chen, Q, Yu, H.L, Xu, J.H. | | Deposit date: | 2017-11-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | Attenuated substrate inhibition of a haloketone reductase via structure-guided loop engineering.
J.Biotechnol., 308, 2020
|
|
