4K85
 
 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 12:0 Ceramide-1-Phosphate (12:0-C1P) | | Descriptor: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-17 | | Release date: | 2013-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4K8N
 
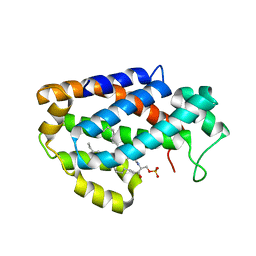 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 18:1 Ceramide-1-Phosphate (18:1-C1P) | | Descriptor: | (2S,3R,4Z)-3-hydroxy-2-[(9E)-octadec-9-enoylamino]octadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-18 | | Release date: | 2013-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4KF6
 
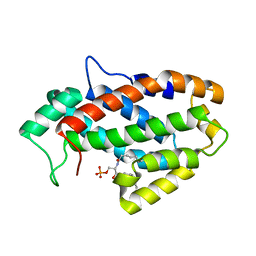 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 8:0 Ceramide-1-Phosphate (8:0-C1P) | | Descriptor: | (2S,3R,4E)-3-hydroxy-2-(octanoylamino)octadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-26 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.195 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4K99
 
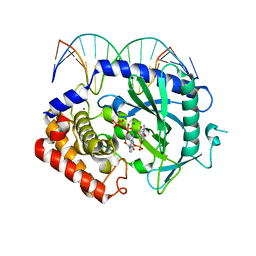 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppdG(2 ,5 )pdG | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, 3'-deoxy-guanosine 5'-monophosphate, Cyclic GMP-AMP synthase, ... | | Authors: | Gao, P, Wu, Y, Patel, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Cell(Cambridge,Mass.), 153, 2013
|
|
8T8E
 
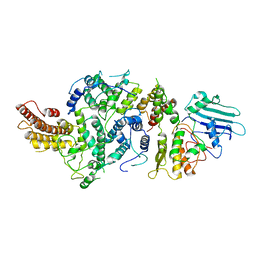 | | cryoEM structure of Smc5/6 5mer | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 6 | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-22 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for Nse5-6 mediated regulation of Smc5/6 functions.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8T8F
 
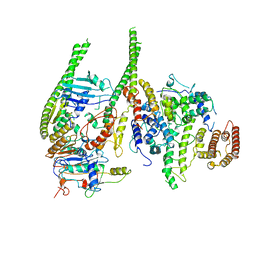 | | Smc5/6 8mer | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 4, Non-structural maintenance of chromosome element 5, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-22 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular basis for Nse5-6 mediated regulation of Smc5/6 functions.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8T66
 
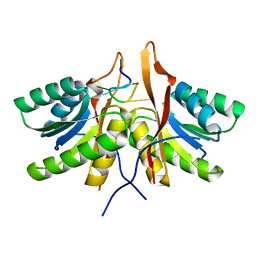 | | cA6 bound Cam1 | | Descriptor: | Cam1, RNA (5'-R(P*AP*AP*AP*AP*A)-3') | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
8T65
 
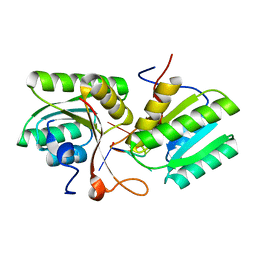 | | cA4 bound Cam1 | | Descriptor: | Cam1, RNA (5'-R(P*AP*AP*AP*A)-3') | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
8T64
 
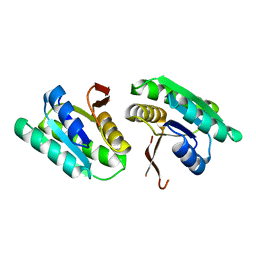 | | Apo Cam1(42-206) | | Descriptor: | Cam1 | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
8URQ
 
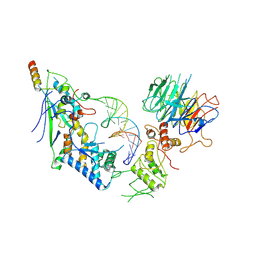 | | Spo11 core complex with gapped DNA | | Descriptor: | Antiviral protein SKI8, MAGNESIUM ION, Meiosis-specific protein SPO11, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the Spo11 core complex bound to DNA.
Nat.Struct.Mol.Biol., 2024
|
|
8URU
 
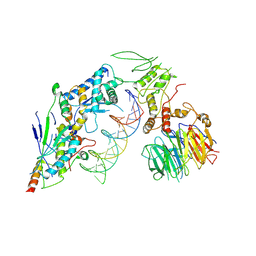 | | Spo11 core complex with hairpin DNA | | Descriptor: | Antiviral protein SKI8, Hairpin DNA, MAGNESIUM ION, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of the Spo11 core complex bound to DNA.
Nat.Struct.Mol.Biol., 2024
|
|
4NJ5
 
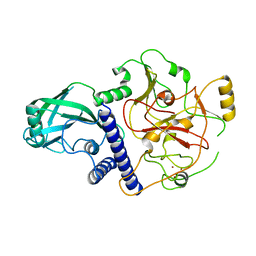 | | Crystal structure of SUVH9 | | Descriptor: | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH9, ZINC ION | | Authors: | Du, J, Patel, D.J. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation.
Nature, 507, 2014
|
|
7DMQ
 
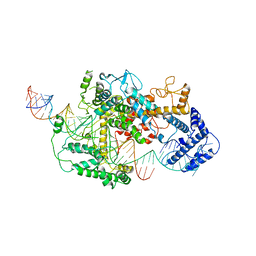 | | Cryo-EM structure of LshCas13a-crRNA-anti-tag RNA complex | | Descriptor: | Anti-tag target RNA, CRISPR RNA, CRISPR/Cas system Cas13a | | Authors: | Wang, B, Zhang, T, Ding, J, Patel, D.J, Yang, H. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for self-cleavage prevention by tag:anti-tag pairing complementarity in type VI Cas13 CRISPR systems.
Mol.Cell, 81, 2021
|
|
7JW2
 
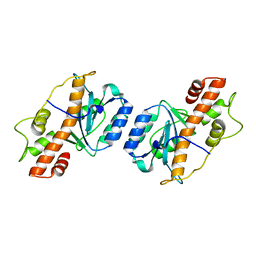 | | Crystal structure of Aedes aegypti Nibbler EXO domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7JW6
 
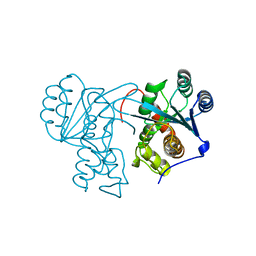 | | Crystal structure of Drosophila Nibbler EXO domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7JW3
 
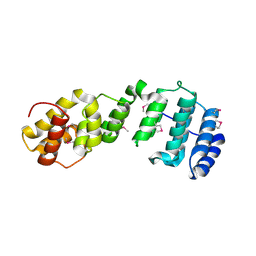 | | Crystal structure of Aedes aegypti Nibbler NTD domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7K3J
 
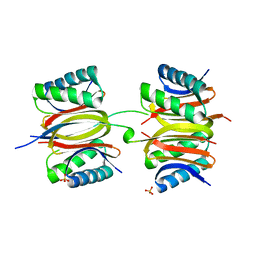 | |
7K3K
 
 | |
7K3L
 
 | |
7TVE
 
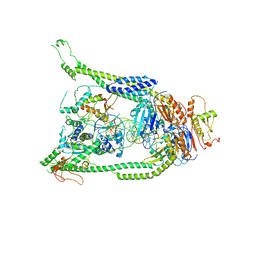 | | ATP and DNA bound SMC5/6 core complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (68-MER), DNA (78-MER), ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2022-02-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of DNA-bound Smc5/6 reveals DNA clamping enabled by multi-subunit conformational changes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4ZNP
 
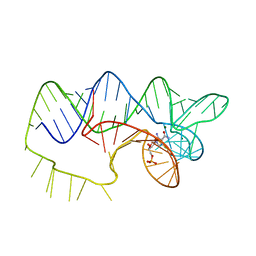 | | The structure of A pfI Riboswitch Bound to ZMP | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, MAGNESIUM ION, pfI Riboswitch | | Authors: | Ren, A, Patel, D.J, Rajashankar, R.K. | | Deposit date: | 2015-05-05 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Global RNA Fold and Molecular Recognition for a pfl Riboswitch Bound to ZMP, a Master Regulator of One-Carbon Metabolism.
Structure, 23, 2015
|
|
4YAZ
 
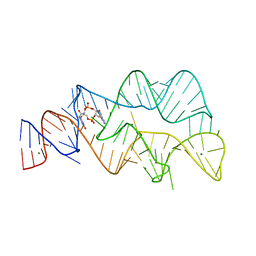 | | 3',3'-cGAMP riboswitch bound with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Ren, A.M, Patel, D.J, Rajashankar, R.K. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Molecular Discrimination by a 3',3'-cGAMP Sensing Riboswitch.
Cell Rep, 11, 2015
|
|
4YB0
 
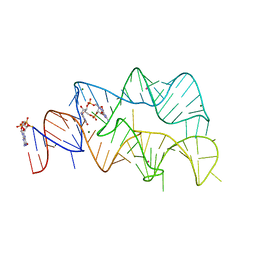 | | 3',3'-cGAMP riboswitch bound with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ren, A.M, Patel, D.J, Rajashankar, R.K. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Structural Basis for Molecular Discrimination by a 3',3'-cGAMP Sensing Riboswitch.
Cell Rep, 11, 2015
|
|
4YB1
 
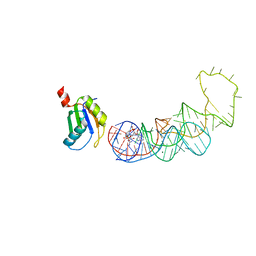 | | 20A Mutant c-di-GMP Vc2 Riboswitch bound with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, MAGNESIUM ION, RNA (91-MER), ... | | Authors: | Ren, A.M, Patel, D.J, Rajashankar, R.K. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Structural Basis for Molecular Discrimination by a 3',3'-cGAMP Sensing Riboswitch.
Cell Rep, 11, 2015
|
|
5Y6O
 
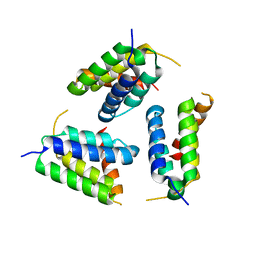 | |
