3MP3
 
 | | Crystal Structure of Human Lyase in complex with inhibitor HG-CoA | | Descriptor: | (3R,5S,9R,21S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9,21-tetrahydroxy-8,8-dimethyl-10,14,19-trioxo-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatricosan-23-oic acid 3,5-dioxide, 3-HYDROXYPENTANEDIOIC ACID, Hydroxymethylglutaryl-CoA lyase, ... | | Authors: | Fu, Z, Runquist, J.A, Montgomery, C, Miziorko, H.M, Kim, J.-J.P. | | Deposit date: | 2010-04-24 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights into human HMG-CoA lyase from structures of Acyl-CoA-containing ternary complexes.
J.Biol.Chem., 285, 2010
|
|
1IVH
 
 | | STRUCTURE OF HUMAN ISOVALERYL-COA DEHYDROGENASE AT 2.6 ANGSTROMS RESOLUTION: STRUCTURAL BASIS FOR SUBSTRATE SPECIFICITY | | Descriptor: | COENZYME A PERSULFIDE, FLAVIN-ADENINE DINUCLEOTIDE, ISOVALERYL-COA DEHYDROGENASE | | Authors: | Tiffany, K.A, Roberts, D.L, Wang, M, Paschke, R, Mohsen, A.-W.A, Vockley, J, Kim, J.J.P. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human isovaleryl-CoA dehydrogenase at 2.6 A resolution: structural basis for substrate specificity,.
Biochemistry, 36, 1997
|
|
1F20
 
 | | CRYSTAL STRUCTURE OF RAT NEURONAL NITRIC-OXIDE SYNTHASE FAD/NADP+ DOMAIN AT 1.9A RESOLUTION. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, GLYCEROL, ... | | Authors: | Zhang, J, Martasek, P, Masters, B.S, Kim, J.P. | | Deposit date: | 2000-05-22 | | Release date: | 2001-10-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the FAD/NADPH-binding domain of rat neuronal nitric-oxide synthase. Comparisons with NADPH-cytochrome P450 oxidoreductase.
J.Biol.Chem., 276, 2001
|
|
2N1H
 
 | |
1EGD
 
 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
1EGC
 
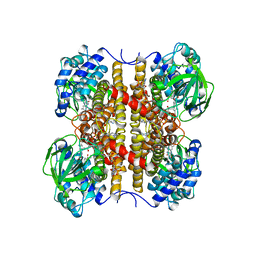 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE COMPLEXED WITH OCTANOYL-COA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE, OCTANOYL-COENZYME A | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
1EGE
 
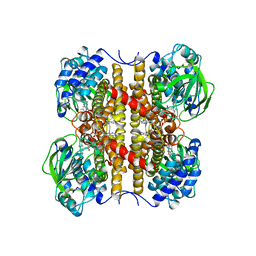 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
2R42
 
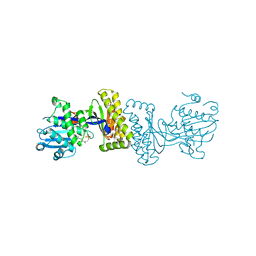 | | The Biochemical and Structural Basis for feedback Inhibition of Mevalonate Kinase and Isoprenoid Metabolism | | Descriptor: | MAGNESIUM ION, Mevalonate kinase, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE | | Authors: | Fu, Z, Voynova, N.E, Miziorko, H.M, Kim, J.P. | | Deposit date: | 2007-08-30 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and Structural Basis for Feedback Inhibition of Mevalonate Kinase and Isoprenoid Metabolism.
Biochemistry, 47, 2008
|
|
2R3V
 
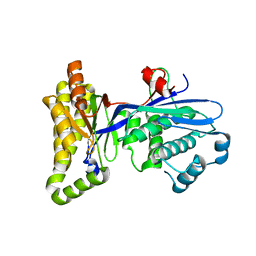 | |
1KEO
 
 | | TWISTS AND TURNS OF THE CD-MPR: LIGAND-BOUND VERSUS LIGAND-FREE RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, cation-dependent mannose-6-phosphate receptor | | Authors: | Olson, L.J, Zhang, J, Dahms, N.M, Kim, J.J. | | Deposit date: | 2001-11-16 | | Release date: | 2002-01-23 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Twists and turns of the cation-dependent mannose 6-phosphate receptor. Ligand-bound versus ligand-free receptor
J.Biol.Chem., 277, 2002
|
|
1BUC
 
 | | THREE-DIMENSIONAL STRUCTURE OF BUTYRYL-COA DEHYDROGENASE FROM MEGASPHAERA ELSDENII | | Descriptor: | ACETOACETYL-COENZYME A, BUTYRYL-COA DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Djordjevic, S, Pace, C.P, Stankovich, M.T, Kim, J.J.P. | | Deposit date: | 1994-09-06 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of butyryl-CoA dehydrogenase from Megasphaera elsdenii.
Biochemistry, 34, 1995
|
|
1JQI
 
 | | Crystal Structure of Rat Short Chain Acyl-CoA Dehydrogenase Complexed With Acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, short chain acyl-CoA dehydrogenase | | Authors: | Battaile, K.P, Molin-Case, J, Paschke, R, Wang, M, Bennett, D, Vockley, J, Kim, J.-J.P. | | Deposit date: | 2001-08-07 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA: comparison with other acyl-CoA dehydrogenases.
J.Biol.Chem., 277, 2002
|
|
1EFV
 
 | |
4XQM
 
 | |
1AMO
 
 | | THREE-DIMENSIONAL STRUCTURE OF NADPH-CYTOCHROME P450 REDUCTASE: PROTOTYPE FOR FMN-AND FAD-CONTAINING ENZYMES | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Wang, M, Roberts, D.L, Paschke, R, Shea, T.M, Masters, B.S.S, Kim, J.J.P. | | Deposit date: | 1997-06-17 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
4YAF
 
 | | rat CYPOR with 2'-AMP | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4Y9U
 
 | | rat CYPOR mutant - G143del | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4Y7C
 
 | | rat CYPOR mutant - G141del/E142N | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-13 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4YAW
 
 | | Reduced CYPOR mutant - G141del | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4YAU
 
 | | Reduced CYPOR mutant - G141del/E142N | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4YAL
 
 | | Reduced CYPOR with 2'-AMP | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4YAO
 
 | | Reduced CYPOR mutant - G143del | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4Y9R
 
 | | rat CYPOR mutant - G141del | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4IQP
 
 | | Crystal Structure of HCRA-W1266A | | Descriptor: | Botulinum neurotoxin type A, GLYCEROL | | Authors: | Fu, Z, Kroken, A.R, Barbieri, J.T, Kim, J.-J.P. | | Deposit date: | 2013-01-12 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhancing the Protective Immune Response against Botulism.
Infect.Immun., 81, 2013
|
|
4FVV
 
 | | Crystal structure of HCR/D-Sa-GBL1/C | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid, Neurotoxin, ... | | Authors: | Fu, Z, Karalewitz, A, Baldwin, M.R, Kim, J.-J.P, Barbieri, J.T. | | Deposit date: | 2012-06-29 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Botulinum neurotoxin serotype C associates with dual ganglioside receptors to facilitate cell entry.
J.Biol.Chem., 287, 2012
|
|
