5EZ4
 
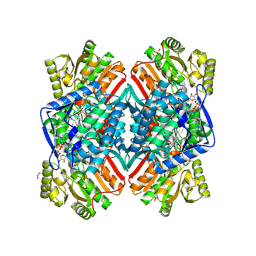 | | 2.11 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-26 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | 2.11 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
5HXM
 
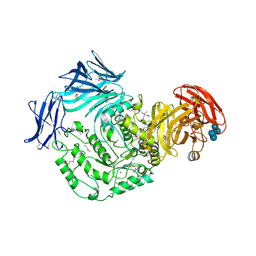 | | Cycloalternan-forming enzyme from Listeria monocytogenes in complex with panose | | Descriptor: | Alpha-xylosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Halavaty, A.S, Light, S.H, Minasov, G, Winsor, J, Grimshaw, S, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-31 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
4R57
 
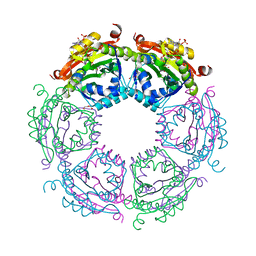 | | Crystal structure of spermidine N-acetyltransferase from Vibrio cholerae in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, DI(HYDROXYETHYL)ETHER, Spermidine n1-acetyltransferase, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
4R9M
 
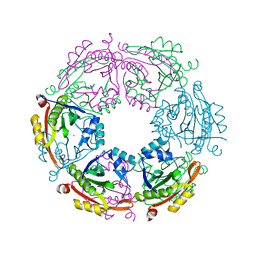 | | Crystal structure of spermidine N-acetyltransferase from Escherichia coli | | Descriptor: | MAGNESIUM ION, Spermidine N(1)-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Grimshaw, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6BI4
 
 | | 2.9 Angstrom Resolution Crystal Structure of dTDP-Glucose 4,6-dehydratase (rfbB) from Bacillus anthracis str. Ames in Complex with NAD. | | Descriptor: | NICKEL (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Halavaty, A.S, Kuhn, M, Shuvalova, L, Minasov, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-31 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure of the Bacillus anthracis dTDP-l-rhamnose biosynthetic pathway enzyme: dTDP-alpha-d-glucose 4,6-dehydratase, RfbB.
J.Struct.Biol., 202, 2018
|
|
6CXD
 
 | | Crystal structure of peptidase B from Yersinia pestis CO92 at 2.75 A resolution | | Descriptor: | Peptidase B, SULFATE ION | | Authors: | Woinska, M, Lipowska, J, Shabalin, I.G, Cymborowski, M, Grimshaw, S, Winsor, J, Shuvalova, L, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-02 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
4JJX
 
 | | Dodecameric structure of spermidine N-acetyltransferase SpeG from Vibrio cholerae O1 biovar eltor | | Descriptor: | Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-08 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
4JHX
 
 | | Crystal structure of anabolic ornithine carbamoyltransferase from Vibrio vulnificus in complex with carbamoylphosphate and arginine | | Descriptor: | ARGININE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Bacal, P, Winsor, J, Grimshaw, S, Grabowski, M, Chordia, M.D, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-05 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
To be Published
|
|
4JLY
 
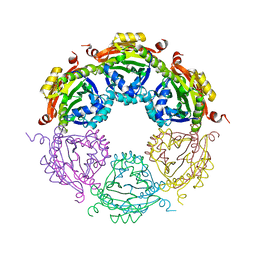 | | Dodecameric structure of spermidine N-acetyltransferase from Vibrio cholerae | | Descriptor: | SULFATE ION, Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
4JFR
 
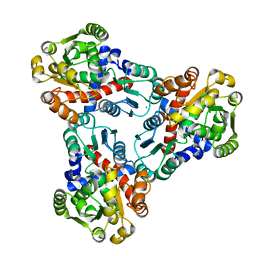 | | Crystal structure of anabolic ornithine carbamoyltransferase from Vibrio vulnificus in complex with carbamoyl phosphate | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Ornithine carbamoyltransferase, ... | | Authors: | Shabalin, I.G, Winsor, J, Grimshaw, S, Osinski, T, Bajor, J, Chordia, M.D, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-28 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
to be published
|
|
4KMQ
 
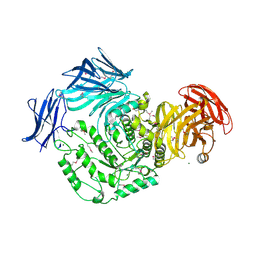 | | 1.9 Angstrom resolution crystal structure of uncharacterized protein lmo2446 from Listeria monocytogenes EGD-e | | Descriptor: | Lmo2446 protein, MAGNESIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Filippova, E.V, Dubrovska, I, Winsor, J, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure to function of an alpha-glucan metabolic pathway that promotes Listeria monocytogenes pathogenesis.
Nat Microbiol, 2, 2016
|
|
4K6A
 
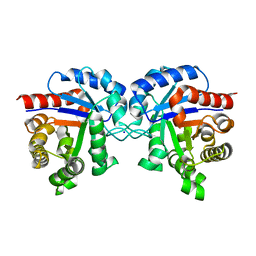 | | Revised Crystal Structure of apo-form of Triosephosphate Isomerase (tpiA) from Escherichia coli at 1.8 Angstrom Resolution. | | Descriptor: | SODIUM ION, Triosephosphate isomerase | | Authors: | Minasov, G, Kuhn, M, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.
PLoS ONE, 9, 2014
|
|
4IUO
 
 | | 1.8 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) K170M Mutant in Complex with Quinate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of schiff base formation.
Biochemistry, 53, 2014
|
|
4EIV
 
 | | 1.37 Angstrom resolution crystal structure of apo-form of a putative deoxyribose-phosphate aldolase from Toxoplasma gondii ME49 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Deoxyribose-phosphate aldolase | | Authors: | Halavaty, A.S, Ruan, J, Minasov, G, Shuvalova, L, Ueno, A, Igarashi, M, Ngo, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-05 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural and Functional Divergence of the Aldolase Fold in Toxoplasma gondii.
J.Mol.Biol., 427, 2015
|
|
4GUH
 
 | | 1.95 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) E86A Mutant in Complex with Dehydroshikimate (Crystal Form #2) | | Descriptor: | (4S,5R)-4,5-dihydroxy-3-oxocyclohex-1-ene-1-carboxylic acid, 3-dehydroquinate dehydratase, NICKEL (II) ION | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Reassessing the type I dehydroquinate dehydratase catalytic triad: Kinetic and structural studies of Glu86 mutants.
Protein Sci., 22, 2013
|
|
4GUI
 
 | | 1.78 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) in Complex with Quinate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, 3-dehydroquinate dehydratase, NICKEL (II) ION | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of schiff base formation.
Biochemistry, 53, 2014
|
|
4GUF
 
 | | 1.5 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) E86A Mutant | | Descriptor: | 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reassessing the type I dehydroquinate dehydratase catalytic triad: Kinetic and structural studies of Glu86 mutants.
Protein Sci., 22, 2013
|
|
4H31
 
 | | Crystal structure of anabolic ornithine carbamoyltransferase from Vibrio vulnificus in complex with carbamoyl phosphate and L-norvaline | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Winsor, J, Grimshaw, S, Osinski, T, Chordia, M.D, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of ornithine carbamoyltransferase from various pathogens
To be Published
|
|
4GUG
 
 | | 1.62 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) E86A Mutant in Complex with Dehydroshikimate (Crystal Form #1) | | Descriptor: | (4S,5R)-4,5-dihydroxy-3-oxocyclohex-1-ene-1-carboxylic acid, 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Reassessing the type I dehydroquinate dehydratase catalytic triad: Kinetic and structural studies of Glu86 mutants.
Protein Sci., 22, 2013
|
|
4GUJ
 
 | | 1.50 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) in Complex with Shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-dehydroquinate dehydratase, ZINC ION | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of schiff base formation.
Biochemistry, 53, 2014
|
|
4MI4
 
 | | Crystal structure of spermidine N-acetyltransferase from Vibrio cholerae in complex with spermine | | Descriptor: | SPERMINE, Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-30 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
5TTA
 
 | | A 1.85A X-Ray Structure from Peptoclostridium difficile 630 of a Hypothetical Protein | | Descriptor: | Putative exported protein | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Cordona-Correa, A, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-02 | | Release date: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A 1.85A X-Ray Structure from Peptoclostridium difficile 630 of a Hypothetical Protein
To Be Published
|
|
4MVJ
 
 | | 2.85 Angstrom Resolution Crystal Structure of Glyceraldehyde 3-phosphate Dehydrogenase A (gapA) from Escherichia coli Modified by Acetyl Phosphate. | | Descriptor: | ACETATE ION, ACETYLPHOSPHATE, CHLORIDE ION, ... | | Authors: | Minasov, G, Kuhn, M, Dubrovska, I, Winsor, J, Shuvalova, L, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.
Plos One, 9, 2014
|
|
4MVA
 
 | | 1.43 Angstrom Resolution Crystal Structure of Triosephosphate Isomerase (tpiA) from Escherichia coli in Complex with Acetyl Phosphate. | | Descriptor: | 1,2-ETHANEDIOL, ACETYLPHOSPHATE, CHLORIDE ION, ... | | Authors: | Minasov, G, Kuhn, M.L, Dubrovska, I, Winsor, J, Shuvalova, L, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.
Plos One, 9, 2014
|
|
3OTR
 
 | | 2.75 Angstrom Crystal Structure of Enolase 1 from Toxoplasma gondii | | Descriptor: | CHLORIDE ION, Enolase, SULFATE ION | | Authors: | Minasov, G, Ruan, J, Shuvalova, L, Halavaty, A, Ngo, H, Tomavo, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-13 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure of bradyzoite-specific enolase from Toxoplasma gondii reveals insights into its dual cytoplasmic and nuclear functions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
