5F38
 
 | | X-ray crystal structure of a thiolase from Escherichia coli at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Acetyl-CoA acetyltransferase, COENZYME A, ... | | Authors: | Ithayaraja, M, Neelanjana, J, Wierenga, R, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2015-12-02 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a thiolase from Escherichia coli at 1.8 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
8R9W
 
 | |
8R9Y
 
 | |
8R9Z
 
 | |
8R9X
 
 | |
8QQK
 
 | | Cryo-EM structure of E. coli cytochrome bo3 quinol oxidase assembled in peptidiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, ... | | Authors: | Gao, Y, Zhang, Y, Hakke, S, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2023-10-05 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of cytochrome bo 3 quinol oxidase assembled in peptidiscs reveals an "open" conformation for potential ubiquinone-8 release.
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
3BP2
 
 | |
1HVE
 
 | | STRUCTURAL AND ELECTROPHYSIOLOGICAL ANALYSIS OF ANNEXIN V MUTANTS. MUTAGENESIS OF HUMAN ANNEXIN V, AN IN VITRO VOLTAGE-GATED CALCIUM CHANNEL, PROVIDES INFORMATION ABOUT THE STRUCTURAL FEATURES OF THE ION PATHWAY, THE VOLTAGE SENSOR AND THE ION SELECTIVITY FILTER | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Burger, A, Huber, R. | | Deposit date: | 1994-06-29 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and electrophysiological analysis of annexin V mutants. Mutagenesis of human annexin V, an in vitro voltage-gated calcium channel, provides information about the structural features of the ion pathway, the voltage sensor and the ion selectivity filter
J.Mol.Biol., 237, 1994
|
|
4OXF
 
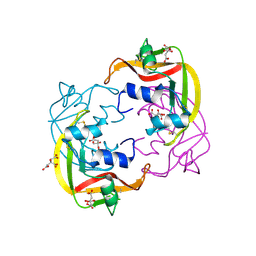 | |
3UQD
 
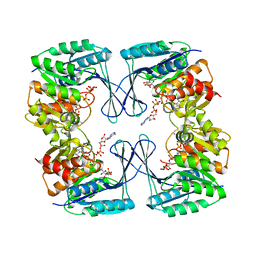 | | Crystal structure of the Phosphofructokinase-2 from Escherichia coli in complex with substrates and products | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructokinase isozyme 2, ... | | Authors: | Pereira, H.M, Caniuguir, A, Baez, M, Cabrera, R, Babul, J. | | Deposit date: | 2011-11-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Studying the phosphoryl transfer mechanism of theE. coliphosphofructokinase-2: from X-ray structure to quantum mechanics/molecular mechanics simulations.
Chem Sci, 10, 2019
|
|
1HVD
 
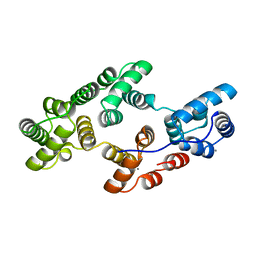 | | STRUCTURAL AND ELECTROPHYSIOLOGICAL ANALYSIS OF ANNEXIN V MUTANTS. MUTAGENESIS OF HUMAN ANNEXIN V, AN IN VITRO VOLTAGE-GATED CALCIUM CHANNEL, PROVIDES INFORMATION ABOUT THE STRUCTURAL FEATURES OF THE ION PATHWAY, THE VOLTAGE SENSOR AND THE ION SELECTIVITY FILTER | | Descriptor: | ANNEXIN V, CALCIUM ION | | Authors: | Burger, A, Huber, R. | | Deposit date: | 1994-06-29 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and electrophysiological analysis of annexin V mutants. Mutagenesis of human annexin V, an in vitro voltage-gated calcium channel, provides information about the structural features of the ion pathway, the voltage sensor and the ion selectivity filter
J.Mol.Biol., 237, 1994
|
|
1HVF
 
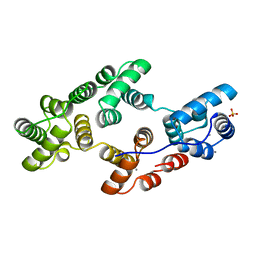 | | STRUCTURAL AND ELECTROPHYSIOLOGICAL ANALYSIS OF ANNEXIN V MUTANTS. MUTAGENESIS OF HUMAN ANNEXIN V, AN IN VITRO VOLTAGE-GATED CALCIUM CHANNEL, PROVIDES INFORMATION ABOUT THE STRUCTURAL FEATURES OF THE ION PATHWAY, THE VOLTAGE SENSOR AND THE ION SELECTIVITY FILTER | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Burger, A, Huber, R. | | Deposit date: | 1994-06-29 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and electrophysiological analysis of annexin V mutants. Mutagenesis of human annexin V, an in vitro voltage-gated calcium channel, provides information about the structural features of the ion pathway, the voltage sensor and the ion selectivity filter
J.Mol.Biol., 237, 1994
|
|
1HVG
 
 | | STRUCTURAL AND ELECTROPHYSIOLOGICAL ANALYSIS OF ANNEXIN V MUTANTS. MUTAGENESIS OF HUMAN ANNEXIN V, AN IN VITRO VOLTAGE-GATED CALCIUM CHANNEL, PROVIDES INFORMATION ABOUT THE STRUCTURAL FEATURES OF THE ION PATHWAY, THE VOLTAGE SENSOR AND THE ION SELECTIVITY FILTER | | Descriptor: | ANNEXIN V | | Authors: | Burger, A, Huber, R. | | Deposit date: | 1994-06-29 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and electrophysiological analysis of annexin V mutants. Mutagenesis of human annexin V, an in vitro voltage-gated calcium channel, provides information about the structural features of the ion pathway, the voltage sensor and the ion selectivity filter
J.Mol.Biol., 237, 1994
|
|
1E94
 
 | | HslV-HslU from E.coli | | Descriptor: | HEAT SHOCK PROTEIN HSLU, HEAT SHOCK PROTEIN HSLV, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Song, H.K, Hartmann, C, Ravishankar, R, Bochtler, M. | | Deposit date: | 2000-10-07 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutational Studies on Hslu and its Docking Mode with Hslv
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4ZJD
 
 | |
4ZJA
 
 | |
4ZJ9
 
 | |
3H5Z
 
 | | Crystal Structure of Leishmania major N-myristoyltransferase with bound myristoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, ... | | Authors: | Qiu, W, Hutchinson, A, Lin, Y.-H, Wernimont, A, Mackenzie, F, Ravichandran, M, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Fairlamb, A.H, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | N-myristoyltransferase inhibitors as new leads to treat sleeping sickness.
Nature, 464, 2010
|
|
1HT2
 
 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-27 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
4FQ9
 
 | | Crystal Structure of 3-hydroxydecanoyl-Acyl Carrier Protein Dehydratase (FabA) from Pseudomonas aeruginosa | | Descriptor: | 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase, GLYCEROL, PHOSPHATE ION | | Authors: | Moynie, L, Mcmahon, S.A, Duthie, F.G, Naismith, J.H. | | Deposit date: | 2012-06-25 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural insights into the mechanism and inhibition of the beta-hydroxydecanoyl-acyl carrier protein dehydratase from Pseudomonas aeruginosa
J.Mol.Biol., 425, 2013
|
|
4ZRC
 
 | | Crystal structure of MSM-13, a putative T1-like thiolase from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase | | Authors: | Janardan, N, Harijan, R.K, Keima, T.R, Wierenga, R, Murthy, M.R.N. | | Deposit date: | 2015-05-12 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Z07
 
 | | Co-crystal structure of the tandem CNB (CNB-A/B) domains of human PKG I beta with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Kim, J.J, Reger, A.S, Arold, S.T, Kim, C. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of PKG I:cGMP Complex Reveals a cGMP-Mediated Dimeric Interface that Facilitates cGMP-Induced Activation.
Structure, 24, 2016
|
|
5BJT
 
 | |
1HT1
 
 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-27 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
7KDT
 
 | |
