2G1O
 
 | |
2G1N
 
 | | Ketopiperazine-based renin inhibitors: Optimization of the "C" ring | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-{2-[6-(2,4-DIAMINO-6-ETHYLPYRIMIDIN-5-YL)-2,2-DIMETHYL-3-OXO-2,3-DIHYDRO-4H-1,4-BENZOTHIAZIN-4-YL]ETHYL}ACETAMIDE, Renin | | Authors: | Holsworth, D.D. | | Deposit date: | 2006-02-14 | | Release date: | 2006-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ketopiperazine-based renin inhibitors: Optimization of the "C" ring
BIOORG.MED.CHEM.LETT., 16, 2006
|
|
5CAL
 
 | | EGFR kinase domain mutant "TMLR" with compound 24 | | Descriptor: | 2,2-dimethyl-3-[(4-{[2-methyl-1-(propan-2-yl)-1H-imidazo[4,5-c]pyridin-6-yl]amino}pyrimidin-2-yl)amino]propanamide, Epidermal growth factor receptor | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
5C8N
 
 | | EGFR kinase domain mutant "TMLR" with compound 23 | | Descriptor: | Epidermal growth factor receptor, N-{2-[4-(2-aminoethyl)-4-methoxypiperidin-1-yl]pyrimidin-4-yl}-2-methyl-1-(propan-2-yl)-1H-imidazo[4,5-c]pyridin-6-amine, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
8T1P
 
 | |
8SZC
 
 | |
8T3K
 
 | |
4OVV
 
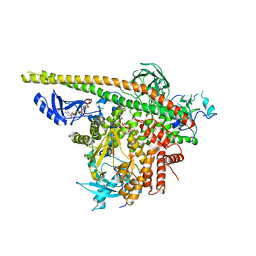 | | Crystal Structure of PI3Kalpha in complex with diC4-PIP2 | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, PHOSPHATE ION, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M, Amzel, L.M. | | Deposit date: | 2014-01-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of nSH2 regulation and lipid binding in PI3K alpha.
Oncotarget, 5, 2014
|
|
5FV5
 
 | | KpFlo11 presents a novel member of the Flo11 family with a unique recognition pattern for homophilic interactions | | Descriptor: | ACETATE ION, Flocculation protein FLO11 | | Authors: | Kraushaar, T, Brueckner, S, Mikolaiski, M, Schreiner, F, Veelders, M, Moesch, H.U, Essen, L.O. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Kin discrimination in social yeast is mediated by cell surface receptors of the Flo11 adhesin family.
Elife, 9, 2020
|
|
5FV6
 
 | | KpFlo11 presents a novel member of the Flo11 family with a unique recognition pattern for homophilic interactions | | Descriptor: | ACETATE ION, Flocculation protein FLO11, GLYCEROL, ... | | Authors: | Kraushaar, T, Brueckner, S, Mikolaiski, M, Schreiner, F, Veelders, M, Moesch, H.U, Essen, L.O. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kin discrimination in social yeast is mediated by cell surface receptors of the Flo11 adhesin family.
Elife, 9, 2020
|
|
5NPL
 
 | | Crystal structure of hexameric CBS-CP12 protein from bloom-forming cyanobacteria, Yb-derivative at 2.8 A resolution | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, Similar to tr|Q8YYT1|Q8YYT1, YTTERBIUM (III) ION | | Authors: | Hackenberg, C, Hakanpaa, J, Antonyuk, S.V, Dittmann, E, Lamzin, V.S. | | Deposit date: | 2017-04-17 | | Release date: | 2018-05-30 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5NMU
 
 | | Structure of hexameric CBS-CP12 protein from bloom-forming cyanobacteria | | Descriptor: | CBS-CP12, CHLORIDE ION | | Authors: | Hackenberg, C, Hakanpaa, J, Antonyuk, S.V, Dittmann, E, Lamzin, V.S. | | Deposit date: | 2017-04-07 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CKT
 
 | | Crystal structure of 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase from Legionella pneumophila Philadelphia 1 | | Descriptor: | 1,2-ETHANEDIOL, 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase from Legionella pneumophila Philadelphia 1
to be published
|
|
6CTE
 
 | | 77Se-NMR probes the protein environment of selenomethionine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Chen, Q, Rozovsky, S. | | Deposit date: | 2018-03-22 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
6C9O
 
 | |
2GJ4
 
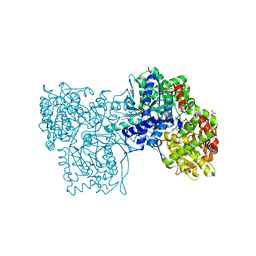 | | Structure of rabbit muscle glycogen phosphorylase in complex with ligand | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 2-CHLORO-N-[(1R,2R)-1-HYDROXY-2,3-DIHYDRO-1H-INDEN-2-YL]-6H-THIENO[2,3-B]PYRROLE-5-CARBOXAMIDE, Glycogen phosphorylase, ... | | Authors: | Otterbein, L.R, Pannifer, A.D, Tucker, J, Breed, J, Oikonomakos, N.G, Rowsell, S, Pauptit, R.A, Claire, M. | | Deposit date: | 2006-03-30 | | Release date: | 2007-02-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel thienopyrrole glycogen phosphorylase inhibitors: synthesis, in vitro SAR and crystallographic studies.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
6CPZ
 
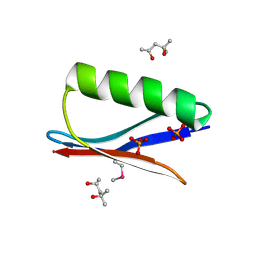 | | Selenomethionine mutant (I6Sem) of protein GB1 examined by X-ray diffraction | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Immunoglobulin G-binding protein G, ... | | Authors: | Chen, Q, Rozovsky, S. | | Deposit date: | 2018-03-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
6CHE
 
 | |
6HSJ
 
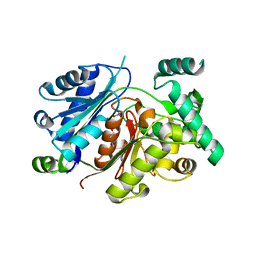 | | Crystal structure of the zebrafish peroxisomal SCP2-thiolase (type-1) in complex with CoA | | Descriptor: | ACETATE ION, COENZYME A, GLYCEROL, ... | | Authors: | Wierenga, R.K, Kiema, T.R, Thapa, C.J. | | Deposit date: | 2018-10-01 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The peroxisomal zebrafish SCP2-thiolase (type-1) is a weak transient dimer as revealed by crystal structures and native mass spectrometry.
Biochem. J., 476, 2019
|
|
6HSP
 
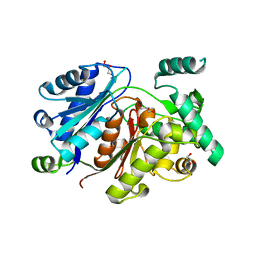 | | Crystal structure of the zebrafish peroxisomal SCP2-thiolase (type-1) in complex with CoA and octanoyl-CoA | | Descriptor: | COENZYME A, GLYCEROL, OCTANOYL-COENZYME A, ... | | Authors: | Wierenga, R.K, Kiema, T.R, Thapa, C.J. | | Deposit date: | 2018-10-01 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The peroxisomal zebrafish SCP2-thiolase (type-1) is a weak transient dimer as revealed by crystal structures and native mass spectrometry.
Biochem. J., 476, 2019
|
|
7AI3
 
 | |
7AI2
 
 | |
6HRV
 
 | |
4HEE
 
 | | Crystal structure of PPARgamma in complex with compound 13 | | Descriptor: | 5-benzyl-2-ethyl-3-{(1S)-5-[2-(1H-tetrazol-5-yl)phenyl]-2,3-dihydro-1H-inden-1-yl}-3,5-dihydro-4H-imidazo[4,5-c]pyridin-4-one, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Han, S. | | Deposit date: | 2012-10-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis, and evaluation of imidazo[4,5-c]pyridin-4-one derivatives with dual activity at angiotensin II type 1 receptor and peroxisome proliferator-activated receptor-gamma
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4OVU
 
 | | Crystal Structure of p110alpha in complex with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2014-01-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural basis of nSH2 regulation and lipid binding in PI3K alpha.
Oncotarget, 5, 2014
|
|
