6IUT
 
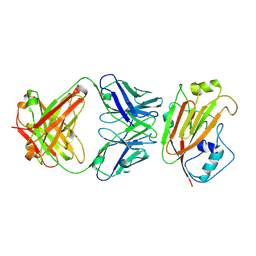 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody AVFluIgG01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, AVFluIgG01 Heavy Chain, ... | | Authors: | Wang, P, Zuo, Y, Sun, J, Zhang, L, Wang, X. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional definition of a vulnerable site on the hemagglutinin of highly pathogenic avian influenza A virus H5N1.
J. Biol. Chem., 294, 2019
|
|
7FJN
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with two T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7FJS
 
 | | Crystal structure of T6 Fab bound to theSARS-CoV-2 RBD of B.1.351 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
3E66
 
 | | Crystal structure of the beta-finger domain of yeast Prp8 | | Descriptor: | PRP8 | | Authors: | Yang, K, Zhang, L, Xu, T, Heroux, A, Zhao, R. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the beta-finger domain of Prp8 reveals analogy to ribosomal proteins.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7FJO
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with three T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7E6G
 
 | | Crystal structure of diguanylate cyclase SiaD in complex with its activator SiaC from Pseudomonas aeruginosa | | Descriptor: | DUF1987 domain-containing protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for diguanylate cyclase activation by its binding partner in Pseudomonas aeruginosa .
Elife, 10, 2021
|
|
7FH3
 
 | | Crystal structure of the ATP sulfurylase domain of human PAPSS2 | | Descriptor: | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthase 2, SULFATE ION, beta-D-glucopyranose | | Authors: | Zhang, P, Zhang, L, Zhang, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the substrate recognition mechanism of ATP-sulfurylase domain of human PAPS synthase 2.
Biochem.Biophys.Res.Commun., 586, 2022
|
|
7FHA
 
 | | Crystal structure of the ATP sulfurylase domain of human PAPSS2 in complex with APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthase 2, POTASSIUM ION, ... | | Authors: | Zhang, P, Zhang, L, Zhang, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the substrate recognition mechanism of ATP-sulfurylase domain of human PAPS synthase 2.
Biochem.Biophys.Res.Commun., 586, 2022
|
|
3PLA
 
 | | Crystal structure of a catalytically active substrate-bound box C/D RNP from Sulfolobus solfataricus | | Descriptor: | 50S ribosomal protein L7Ae, C/D guide RNA, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, ... | | Authors: | Lin, J, Lai, S, Jia, R, Xu, A, Zhang, L, Lu, J, Ye, K. | | Deposit date: | 2010-11-15 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for site-specific ribose methylation by box C/D RNA protein complexes.
Nature, 469, 2011
|
|
3TBO
 
 | |
8IF8
 
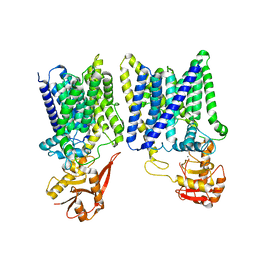 | | Arabinosyltransferase AftA | | Descriptor: | CALCIUM ION, Galactan 5-O-arabinofuranosyltransferase | | Authors: | Gong, Y.C, Rao, Z.H, Zhang, L. | | Deposit date: | 2023-02-17 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the priming arabinosyltransferase AftA required for AG biosynthesis of Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3TBN
 
 | |
3TBM
 
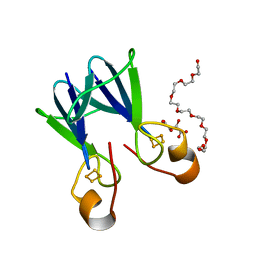 | | Crystal structure of a type 4 CDGSH iron-sulfur protein. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, L(+)-TARTARIC ACID, NONAETHYLENE GLYCOL, ... | | Authors: | Lin, J, Zhang, L, Ye, K. | | Deposit date: | 2011-08-07 | | Release date: | 2011-10-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure and Molecular Evolution of CDGSH Iron-Sulfur Domains.
Plos One, 6, 2011
|
|
7WUQ
 
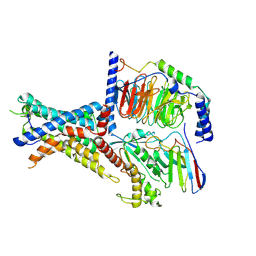 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | Descriptor: | Adhesion G-protein coupled receptor G2,mCherry, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Q.T, Guo, S.C, Xiao, P, Sun, J.P, Yu, X, Gou, L, Kong, L.L, Zhang, L. | | Deposit date: | 2022-02-09 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
7VWP
 
 | | Structure of the flavin-dependent monooxygenase FlsO1 from the biosynthesis of fluostatinsin | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FlsO1, PHOSPHATE ION, ... | | Authors: | Zhang, Y, Yang, C, Zhang, L, Zhang, C. | | Deposit date: | 2021-11-11 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural insights of multifunctional flavin-dependent monooxygenase FlsO1-catalyzed unexpected xanthone formation
Nat Commun, 13, 2022
|
|
6J5I
 
 | | Cryo-EM structure of the mammalian DP-state ATP synthase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F1 subunit epsilon, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
7EBS
 
 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT from silkworm | | Descriptor: | Juvenile hormone acid methyltransferase | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, L, Xu, H.Y, Xia, Q.Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for juvenile hormone biosynthesis by the juvenile hormone acid methyltransferase.
J.Biol.Chem., 297, 2021
|
|
7EBX
 
 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT in complex with S-adenosyl-L-homocysteine. | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, juvenile hormone acid methyltransferase | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, L, Xu, H.Y, Xia, Q.Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural basis for juvenile hormone biosynthesis by the juvenile hormone acid methyltransferase.
J.Biol.Chem., 297, 2021
|
|
7EC0
 
 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT in complex with S-Adenosyl homocysteine and methyl farnesoate | | Descriptor: | Juvenile hormone acid methyltransferase, Methyl farnesoate, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, L, Xu, H.Y, Xia, Q.Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Structural basis for juvenile hormone biosynthesis by the juvenile hormone acid methyltransferase.
J.Biol.Chem., 297, 2021
|
|
6J54
 
 | | Cryo-EM structure of the mammalian E-state ATP synthase FO section | | Descriptor: | ATP synthase membrane subunit 6.8PL, ATP synthase membrane subunit DAPIT, ATP synthase peripheral stalk-membrane subunit b, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
6J5A
 
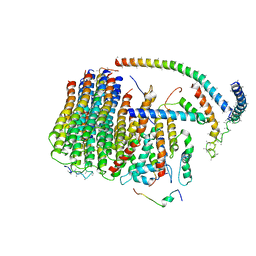 | | Cryo-EM structure of the mammalian DP-state ATP synthase FO section | | Descriptor: | ATP synthase membrane subunit 6.8PL, ATP synthase membrane subunit DAPIT, ATP synthase peripheral stalk-membrane subunit b, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
6IZO
 
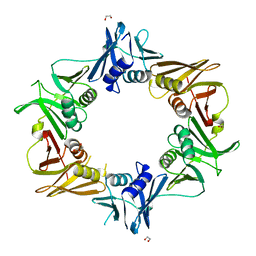 | | Crystal structure of DNA polymerase sliding clamp from Caulobacter crescentus | | Descriptor: | 1,2-ETHANEDIOL, Beta sliding clamp, DI(HYDROXYETHYL)ETHER | | Authors: | Jiang, X, Zhang, L, Teng, M, Li, X. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Caulobacter crescentus beta sliding clamp employs a noncanonical regulatory model of DNA replication.
Febs J., 287, 2020
|
|
7W5P
 
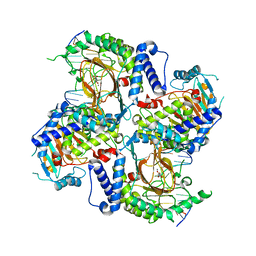 | | Crystal Structure of the dioxygenase CcTet from Coprinopsis cinereain bound to 12bp N6-methyldeoxyadenine (6mA) containing duplex DNA | | Descriptor: | CcTet, DNA, DNA (12-MER), ... | | Authors: | Mu, Y.J, Zhang, L, Zhang, L. | | Deposit date: | 2021-11-30 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A fungal dioxygenase CcTet serves as a eukaryotic 6mA demethylase on duplex DNA.
Nat.Chem.Biol., 18, 2022
|
|
6J5K
 
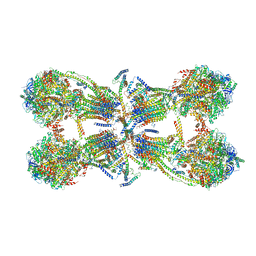 | | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F1 subunit alpha, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
7EVP
 
 | | Cryo-EM structure of the Gp168-beta-clamp complex | | Descriptor: | Beta sliding clamp, Sliding clamp inhibitor | | Authors: | Liu, B, Li, S, Liu, Y, Chen, H, Hu, Z, Wang, Z, Gou, L, Zhang, L, Ma, B, Wang, H, Matthews, S, Wang, Y, Zhang, K. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage Twort protein Gp168 is a beta-clamp inhibitor by occupying the DNA sliding channel.
Nucleic Acids Res., 49, 2021
|
|
