5S6X
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z2889976755 | | Descriptor: | 1-(2,4-dimethyl-1H-imidazol-5-yl)methanamine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S70
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-181428 | | Descriptor: | (5R)-2-methyl-4,5,6,7-tetrahydro-1H-benzimidazol-5-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAF
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-321461 | | Descriptor: | 6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAI
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z1424343998 | | Descriptor: | N-{2-[(propan-2-yl)sulfanyl]phenyl}urea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAC
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z59181945 | | Descriptor: | CITRIC ACID, N-hydroxyquinoline-2-carboxamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SA9
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z2697514548 | | Descriptor: | 1-methylindazole-3-carboxamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SA5
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z1530301542 | | Descriptor: | 4-ethyl-2-(1H-imidazol-5-yl)-1,3-thiazole, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SA7
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z1673618163 | | Descriptor: | 4-amino-N-(2-hydroxyethyl)-N-methylbenzene-1-sulfonamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAG
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-1605072 | | Descriptor: | 3-(1H-imidazol-2-yl)propan-1-amine, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SA6
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z2856434783 | | Descriptor: | N-benzyl-1-(4-fluorophenyl)methanamine, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.517 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAB
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z31504642 | | Descriptor: | 2-methoxy-N-phenylacetamide, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAE
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z3219959731 | | Descriptor: | CITRIC ACID, Uridylate-specific endoribonuclease, pyridazin-3(2H)-one | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAA
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z319891284 | | Descriptor: | 3-[(2S)-1-(methanesulfonyl)pyrrolidin-2-yl]-5-methyl-1,2-oxazole, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.239 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SA4
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z239136710 | | Descriptor: | N-(5-methyl-1H-pyrazol-3-yl)acetamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SA8
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z68299550 | | Descriptor: | 3-chloro-N-(1-hydroxy-2-methylpropan-2-yl)benzamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAH
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-100112 | | Descriptor: | 2-methyl-5,6,7,8-tetrahydropyrido[4,3-c]pyridazin-3(2H)-one, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAD
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z425449682 | | Descriptor: | (3-phenyl-1,2-oxazol-5-yl)methylazanium, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
4MV0
 
 | | IspH in complex with pyridin-2-ylmethyl diphosphate | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, pyridin-2-ylmethyl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Song, Y, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | Deposit date: | 2013-09-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
4MUY
 
 | | IspH in complex with pyridin-4-ylmethyl diphosphate | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, pyridin-4-ylmethyl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Song, Y, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | Deposit date: | 2013-09-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
1PPW
 
 | |
1PPV
 
 | |
3W7F
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from staphylococcus aureus complexed with farnesyl thiopyrophosphate | | Descriptor: | Dehydrosqualene synthase, MAGNESIUM ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H, Oldfield, E. | | Deposit date: | 2013-02-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence.
Science, 319, 2008
|
|
1X84
 
 | | IPP isomerase (wt) reacted with (S)-bromohydrine of IPP | | Descriptor: | (S)-4-BROMO-3-HYDROXY-3-METHYLBUTYL DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION, ... | | Authors: | Wouters, J, Oldfield, E. | | Deposit date: | 2004-08-17 | | Release date: | 2005-01-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Crystallographic Investigation of Phosphoantigen Binding to Isopentenyl Pyrophosphate/Dimethylallyl Pyrophosphate Isomerase
J.Am.Chem.Soc., 127, 2005
|
|
1X83
 
 | | Y104F IPP isomerase reacted with (S)-bromohydrine of IPP | | Descriptor: | (S)-4-BROMO-3-HYDROXY-3-METHYLBUTYL DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION, ... | | Authors: | Wouters, J, Oldfield, E. | | Deposit date: | 2004-08-17 | | Release date: | 2005-01-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Crystallographic Investigation of Phosphoantigen Binding to Isopentenyl Pyrophosphate/Dimethylallyl Pyrophosphate Isomerase
J.Am.Chem.Soc., 127, 2005
|
|
2E9C
 
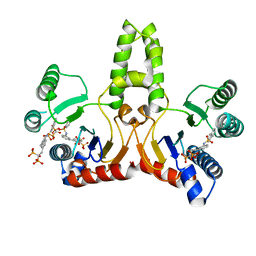 | | E. coli undecaprenyl pyrophosphate synthase in complex with BPH-675 | | Descriptor: | 1-HYDROXY-2-[3'-(NAPHTHALENE-2-SULFONYLAMINO)-BIPHENYL-3-YL]ETHYLIDENE-1,1-BISPHOSPHONIC ACID, Undecaprenyl pyrophosphate synthetase | | Authors: | Guo, R.T, Ko, T.P, Cao, R, Liang, P.H, Oldfield, E, Wang, A.H.J. | | Deposit date: | 2007-01-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
