4ET0
 
 | |
4MNG
 
 | | Structure of the DP10.7 TCR with CD1d-sulfatide | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Luoma, A.M, Adams, E.J. | | Deposit date: | 2013-09-10 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.0058 Å) | | Cite: | Crystal Structure of V delta 1 T Cell Receptor in Complex with CD1d-Sulfatide Shows MHC-like Recognition of a Self-Lipid by Human gamma delta T Cells.
Immunity, 39, 2013
|
|
4MQ7
 
 | | Structure of human CD1d-sulfatide | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Luoma, A.M, Adams, E.J. | | Deposit date: | 2013-09-15 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6032 Å) | | Cite: | Crystal Structure of V delta 1 T Cell Receptor in Complex with CD1d-Sulfatide Shows MHC-like Recognition of a Self-Lipid by Human gamma delta T Cells.
Immunity, 39, 2013
|
|
4MNH
 
 | | Structure of the DP10.7 TCR | | Descriptor: | Human nkt tcr alpha chain, T-cell receptor gamma chain V region PT-gamma-1/2, Human nkt tcr beta chain | | Authors: | Luoma, A.M, Adams, E.J. | | Deposit date: | 2013-09-10 | | Release date: | 2013-12-18 | | Last modified: | 2017-06-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of V delta 1 T Cell Receptor in Complex with CD1d-Sulfatide Shows MHC-like Recognition of a Self-Lipid by Human gamma delta T Cells.
Immunity, 39, 2013
|
|
4NDM
 
 | | Structure of the AB18.1 TCR | | Descriptor: | Human nkt tcr alpha chain, T-cell gamma protein,T-cell receptor beta-2 chain C region | | Authors: | Luoma, A.M, Adams, E.J. | | Deposit date: | 2013-10-27 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.0065 Å) | | Cite: | Crystal Structure of V delta 1 T Cell Receptor in Complex with CD1d-Sulfatide Shows MHC-like Recognition of a Self-Lipid by Human gamma delta T Cells.
Immunity, 39, 2013
|
|
1MYN
 
 | | SOLUTION STRUCTURE OF DROSOMYCIN, THE FIRST INDUCIBLE ANTIFUNGAL PROTEIN FROM INSECTS, NMR, 15 STRUCTURES | | Descriptor: | DROSOMYCIN | | Authors: | Landon, C, Sodano, P, Hetru, C, Hoffmann, J.A, Ptak, M. | | Deposit date: | 1996-12-26 | | Release date: | 1997-12-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of drosomycin, the first inducible antifungal protein from insects.
Protein Sci., 6, 1997
|
|
2GJJ
 
 | | Crystal structure of a single chain antibody scA21 against Her2/ErbB2 | | Descriptor: | A21 single-chain antibody fragment against erbB2, GLYCEROL | | Authors: | Zhu, Z. | | Deposit date: | 2006-03-31 | | Release date: | 2006-10-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Epitope mapping and structural analysis of an anti-ErbB2 antibody A21: Molecular basis for tumor inhibitory mechanism
Proteins, 70, 2007
|
|
4H4B
 
 | | Human cytosolic 5'-nucleotidase II in complex with Anthraquinone-2,6- disulfonic acid | | Descriptor: | 9,10-dioxo-9,10-dihydroanthracene-2,6-disulfonic acid, Cytosolic purine 5'-nucleotidase, GLYCEROL, ... | | Authors: | Rhimi, M, Aghajari, N. | | Deposit date: | 2012-09-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification and characterization of inhibitors of cytoplasmic 5'-nucleotidase cN-II issued from virtual screening.
Biochem Pharmacol, 85, 2013
|
|
2QKN
 
 | |
2QV1
 
 | | Crystal structure of HCV NS3-4A V36M mutant | | Descriptor: | NS3, ZINC ION, peptide | | Authors: | Wei, Y. | | Deposit date: | 2007-08-07 | | Release date: | 2008-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phenotypic and Structural Analyses of HCV NS3 Protease Val36 Variants
To be Published
|
|
3TKJ
 
 | | Crystal Structure of Human Asparaginase-like Protein 1 Thr168Ala | | Descriptor: | L-asparaginase, SODIUM ION, SULFATE ION, ... | | Authors: | Li, W.Z, Yogesha, S.D, Liu, J, Zhang, Y. | | Deposit date: | 2011-08-26 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uncoupling Intramolecular Processing and Substrate Hydrolysis in the N-Terminal Nucleophile Hydrolase hASRGL1 by Circular Permutation.
Acs Chem.Biol., 7, 2012
|
|
3UIZ
 
 | |
2ZKU
 
 | | Structure of hepatitis C virus NS5B polymerase in a new crystal form | | Descriptor: | ACETIC ACID, GLYCEROL, Genome polyprotein | | Authors: | Biswal, B.K. | | Deposit date: | 2008-03-31 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Hepatitis C Virus Ns5B Polymerase in a New Crystal Form: Insights Into Oligomerisation and Allosteric Nucleotide Binding Site
To be Published
|
|
3UIY
 
 | |
1D6B
 
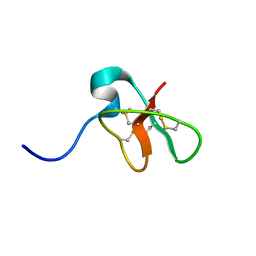 | | SOLUTION STRUCTURE OF DEFENSIN-LIKE PEPTIDE-2 (DLP-2) FROM PLATYPUS VENOM | | Descriptor: | DEFENSIN-LIKE PEPTIDE-2 | | Authors: | Torres, A.M, De Plater, G.M, Doverskog, M, C Birinyi-Strachan, L, Nicholson, G.M, Gallagher, C.H, Kuchel, P.W. | | Deposit date: | 1999-10-12 | | Release date: | 2000-06-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Defensin-like peptide-2 from platypus venom: member of a class of peptides with a distinct structural fold.
Biochem.J., 348, 2000
|
|
2OFU
 
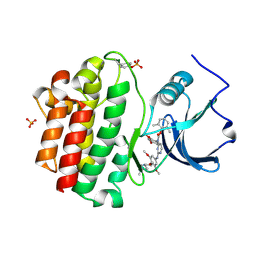 | | x-ray crystal structure of 2-aminopyrimidine carbamate 43 bound to Lck | | Descriptor: | 2,6-DIMETHYLPHENYL 2-(3,5-DIMETHOXY-4-(3-(4-METHYLPIPERAZIN-1-YL)PROPOXY)PHENYLAMINO)PYRIMIDIN- 4-YL(2,4-DIMETHOXYPHENYL)CARBAMATE, Proto-oncogene tyrosine-protein kinase LCK, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2007-01-04 | | Release date: | 2007-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel 2-Aminopyrimidine Carbamates as Potent and Orally Active Inhibitors of Lck: Synthesis, SAR, and in Vivo Antiinflammatory Activity
J.Med.Chem., 49, 2006
|
|
2OFV
 
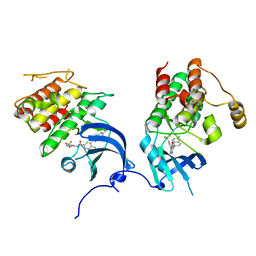 | | crystal structure of aminoquinazoline 1 bound to Lck | | Descriptor: | 3-(2-AMINOQUINAZOLIN-6-YL)-4-METHYL-N-[3-(TRIFLUOROMETHYL)PHENYL]BENZAMIDE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2007-01-04 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Aminoquinazolines as Potent, Orally Bioavailable Inhibitors of Lck: Synthesis, SAR, and in Vivo Anti-Inflammatory Activity
J.Med.Chem., 49, 2006
|
|
2OG8
 
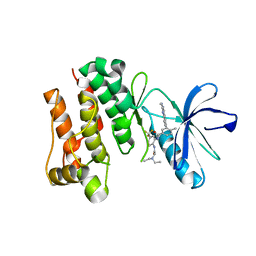 | | crystal structure of aminoquinazoline 36 bound to Lck | | Descriptor: | N-{2-[(N,N-DIETHYLGLYCYL)AMINO]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[2-(METHYLAMINO)QUINAZOLIN-6-YL]BENZAMIDE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Aminoquinazolines as Potent, Orally Bioavailable Inhibitors of Lck: Synthesis, SAR, and in Vivo Anti-Inflammatory Activity
J.Med.Chem., 49, 2006
|
|
4KIO
 
 | | Kinase domain mutant of human Itk in complex with a covalently-binding inhibitor | | Descriptor: | 1-[(3S)-3-{[4-(morpholin-4-ylmethyl)-6-([1,3]thiazolo[5,4-b]pyridin-2-ylamino)pyrimidin-2-yl]amino}pyrrolidin-1-yl]prop-2-en-1-one, 1-[(3S)-3-{[4-(morpholin-4-ylmethyl)-6-([1,3]thiazolo[5,4-b]pyridin-2-ylamino)pyrimidin-2-yl]amino}pyrrolidin-1-yl]propan-1-one, GLYCEROL, ... | | Authors: | Somers, D.O. | | Deposit date: | 2013-05-02 | | Release date: | 2013-08-21 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of novel irreversible inhibitors of interleukin (IL)-2-inducible tyrosine kinase (Itk) by targeting cysteine 442 in the ATP pocket.
J.Biol.Chem., 288, 2013
|
|
3EYC
 
 | | New crystal structure of human tear lipocalin in complex with 1,4-butanediol in space group P21 | | Descriptor: | 1,4-BUTANEDIOL, Lipocalin-1 | | Authors: | Breustedt, D.A, Keil, L, Skerra, A. | | Deposit date: | 2008-10-20 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new crystal form of human tear lipocalin reveals high flexibility in the loop region and induced fit in the ligand cavity
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1MEY
 
 | | CRYSTAL STRUCTURE OF A DESIGNED ZINC FINGER PROTEIN BOUND TO DNA | | Descriptor: | CHLORIDE ION, CONSENSUS ZINC FINGER, DNA (5'-D(*AP*TP*GP*AP*GP*GP*CP*AP*GP*AP*AP*CP*T)-3'), ... | | Authors: | Kim, C.A, Berg, J.M. | | Deposit date: | 1996-09-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A 2.2 A Resolution Crystal Structure of a Designed Zinc Finger Protein Bound to DNA
Nat.Struct.Biol., 3, 1996
|
|
2QPM
 
 | |
3JXS
 
 | | Crystal structure of XG34, an evolved xyloglucan binding CBM | | Descriptor: | ACETATE ION, CALCIUM ION, Xylanase | | Authors: | Divne, C, Tan, T.-C, Brumer, H, Gullfot, F. | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of XG-34, an evolved xyloglucan-specific carbohydrate-binding module.
Proteins, 78, 2009
|
|
3BW7
 
 | |
3C0P
 
 | |
