8T8W
 
 | | Crystal structure of Terrestrivirus Inositol pyrophosphatase kinase in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CADMIUM ION, Kinase, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8T92
 
 | | Crystal structure of Terrestrivirus Inositol pyrophosphatase kinase in complex with ADP and scyllo-D-(1,2,3,4)-IP4 | | Descriptor: | (1S,2R,3R,4S,5R,6R)-5,6-dihydroxycyclohexane-1,2,3,4-tetrayl tetrakis[dihydrogen (phosphate)], ADENOSINE-5'-DIPHOSPHATE, Kinase, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8T90
 
 | | Crystal structure of Terrestrivirus Inositol pyrophosphatase kinase in complex with ADP and myo-(3OH)IP5 | | Descriptor: | (1R,2R,3R,4S,5R,6R)-6-hydroxycyclohexane-1,2,3,4,5-pentayl pentakis[dihydrogen (phosphate)], ADENOSINE-5'-DIPHOSPHATE, Kinase, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8T91
 
 | | Crystal structure of Terrestrivirus Inositol pyrophosphatase kinase in complex with ADP and myo-IP6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Kinase, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8T8Y
 
 | | Crystal structure of Terrestrivirus inositol pyrophosphate kinase in complex with ADP and 1,4,5-InsP3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Kinase, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8T95
 
 | | Crystal structure of Terrestrivirus Inositol pyrophosphatase kinase in complex with ADP and scyllo-(1,2,4,5)-IP4 | | Descriptor: | (1S,3R,4R,6S)-1,3,4,6-TETRAPKISPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Kinase, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8T8U
 
 | | Crystal structure of Terrestrivirus full-length Inositol pyrophosphate kinase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinase, MAGNESIUM ION, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8T8X
 
 | | Crystal structure of Terrestrivirus inositol pyrophosphate kinase in complex with AMP-PNP | | Descriptor: | CADMIUM ION, Kinase, PHOSPHATE ION, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8T8Z
 
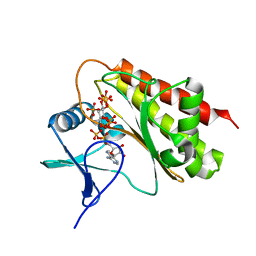 | | Crystal structure of Terrestrivirus Inositol pyrophosphatase kinase in complex with ADP and myo-(1OH)IP5 | | Descriptor: | (1S,2R,3S,4S,5S,6S)-6-hydroxycyclohexane-1,2,3,4,5-pentayl pentakis[dihydrogen (phosphate)], ADENOSINE-5'-DIPHOSPHATE, Kinase, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8TF9
 
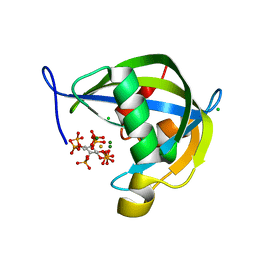 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with myo-5-IP7, produced upon myo-IP6 phosphorylation by TvI | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-07-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8TFA
 
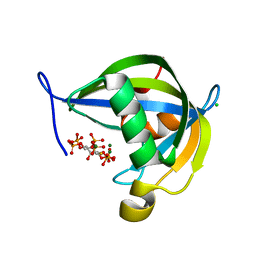 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with myo-5-PP-IP4, produced upon myo-(2OH)-IP5 phosphorylation by TvIPK | | Descriptor: | (1r,2R,3S,4r,5R,6S)-4-hydroxy-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-07-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
5A8H
 
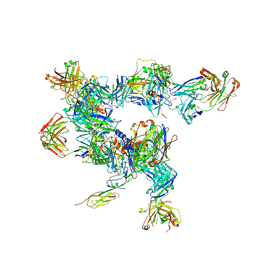 | | cryo-ET subtomogram averaging of BG505 SOSIP.664 in complex with sCD4, 17b, and 8ANC195 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB OF BROADLY NEUTRALIZING ANTIBODY 17B, FAB OF BROADLY NEUTRALIZING ANTIBODY 8ANC195 VARIANT G52K5, ... | | Authors: | Scharf, L, Wang, H, Gao, H, Chen, S, McDowall, A, Bjorkman, P. | | Deposit date: | 2015-07-15 | | Release date: | 2015-08-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Broadly Neutralizing Antibody 8ANC195 Recognizes Closed and Open States of HIV-1 Env.
Cell, 162, 2015
|
|
5A7X
 
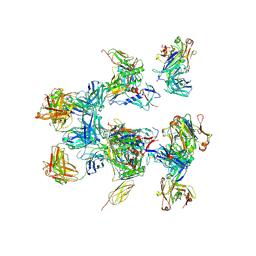 | | negative stain EM of BG505 SOSIP.664 in complex with sCD4, 17b, and 8ANC195 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB OF BROADLY NEUTRALIZING ANTIBODY 17B, FAB OF BROADLY NEUTRALIZING ANTIBODY 8ANC195, ... | | Authors: | Scharf, L, Wang, H, Gao, H, Chen, S, McDowall, A, Bjorkman, P. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Broadly Neutralizing Antibody 8Anc195 Recognizes Closed and Open States of HIV-1 Env.
Cell(Cambridge,Mass.), 162, 2015
|
|
3C4A
 
 | | Crystal structure of vioD hydroxylase in complex with FAD from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CvR158 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable tryptophan hydroxylase vioD | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Wang, H, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-29 | | Release date: | 2008-02-05 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of vioD hydroxylase in complex with FAD from Chromobacterium violaceum.
To be Published
|
|
6DLV
 
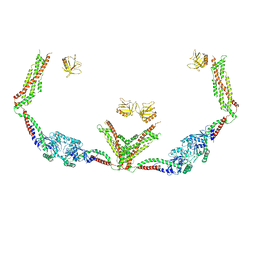 | | Cryo-EM of the GTP-bound human dynamin-1 polymer assembled on the membrane in the super constricted state | | Descriptor: | Dynamin-1 | | Authors: | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | Deposit date: | 2018-06-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
3G3N
 
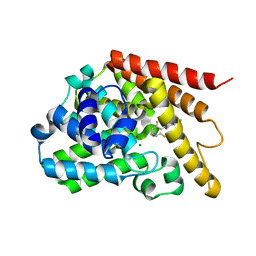 | | PDE7A catalytic domain in complex with 3-(2,6-difluorophenyl)-2-(methylthio)quinazolin-4(3H)-one | | Descriptor: | 3-(2,6-difluorophenyl)-2-(methylthio)quinazolin-4(3H)-one, High affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A, MAGNESIUM ION, ... | | Authors: | Castano, T, Wang, H. | | Deposit date: | 2009-02-02 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis, structural analysis, and biological evaluation of thioxoquinazoline derivatives as phosphodiesterase 7 inhibitors
Chemmedchem, 4, 2009
|
|
8T8M
 
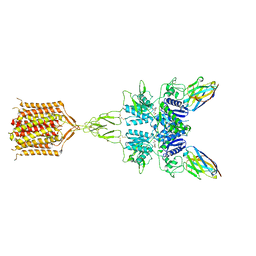 | | Quis-bound intermediate mGlu5 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 5, Nb43 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-22 | | Release date: | 2023-10-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8T6J
 
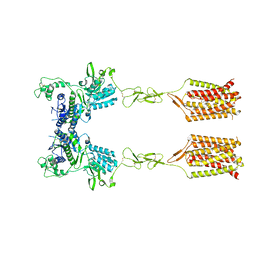 | | CDPPB-bound inactive mGlu5 | | Descriptor: | 3-cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide, Metabotropic glutamate receptor 5 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-16 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8T7H
 
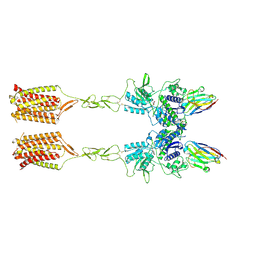 | | Quis-bound intermediate mGlu5 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 5, Nanobody 43 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-20 | | Release date: | 2023-10-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8TAO
 
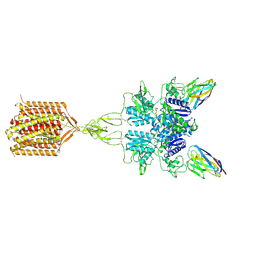 | | Quis and CDPPB bound active mGlu5 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 3-cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide, Metabotropic glutamate receptor 5, ... | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
4TMP
 
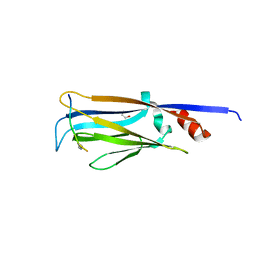 | | Crystal structure of AF9 YEATS bound to H3K9ac peptide | | Descriptor: | 1,2-ETHANEDIOL, ALA-ARG-THR-LYS-GLN-THR-ALA-ARG-ALY-SER-THR, Protein AF-9 | | Authors: | Li, H, Li, Y, Wang, H, Ren, Y. | | Deposit date: | 2014-06-02 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation.
Cell, 159, 2014
|
|
5YIJ
 
 | | Structure of a Legionella effector with substrates | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, SdeA, Ubiquitin | | Authors: | Feng, Y, Mu, Y, Wang, H. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structural basis of ubiquitin modification by the Legionella effector SdeA.
Nature, 557, 2018
|
|
3BJC
 
 | | Crystal structure of the PDE5A catalytic domain in complex with a novel inhibitor | | Descriptor: | 5-ethoxy-4-(1-methyl-7-oxo-3-propyl-6,7-dihydro-1H-pyrazolo[4,3-d]pyrimidin-5-yl)thiophene-2-sulfonamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, G, Wang, H, Howard, R, Cai, J, Wan, Y, Ke, H. | | Deposit date: | 2007-12-03 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An insight into the pharmacophores of phosphodiesterase-5 inhibitors from synthetic and crystal structural studies
BIOCHEM.PHARM., 75, 2008
|
|
1Q2I
 
 | | NMR SOLUTION STRUCTURE OF A PEPTIDE FROM THE MDM-2 BINDING DOMAIN OF THE P53 PROTEIN THAT IS SELECTIVELY CYTOTOXIC TO CANCER CELLS | | Descriptor: | PNC27 | | Authors: | Rosal, R, Pincus, M.R, Brandt-Rauf, P.W, Fine, R.L, Wang, H. | | Deposit date: | 2003-07-24 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a peptide from the mdm-2 binding domain of the p53 protein that is selectively cytotoxic to cancer cells
Biochemistry, 43, 2004
|
|
1Q2F
 
 | | NMR SOLUTION STRUCTURE OF A PEPTIDE FROM THE MDM-2 BINDING DOMAIN OF THE P53 PROTEIN THAT IS SELECTIVELY CYTOTOXIC TO CANCER CELLS | | Descriptor: | PNC27 | | Authors: | Rosal, R, Pincus, M.R, Brandt-Rauf, P.W, Fine, R.L, Wang, H. | | Deposit date: | 2003-07-24 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a peptide from the mdm-2 binding domain of the p53 protein that is selectively cytotoxic to cancer cells
Biochemistry, 43, 2004
|
|
