7ND5
 
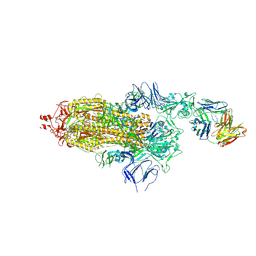 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-150 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-150 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
8QRY
 
 | |
3I3G
 
 | | Crystal Structure of Trypanosoma brucei N-acetyltransferase (Tb11.01.2886) at 1.86A | | Descriptor: | N-acetyltransferase | | Authors: | Qiu, W, Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, Mackenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, J Ferguson, M.A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure Trypanosoma brucei N-acetyltransferase (Tb11.01.2886) at 1.86A
To be Published
|
|
7NDD
 
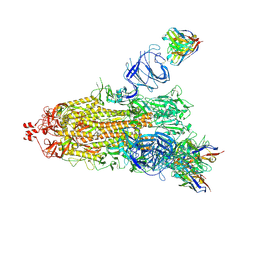 | | EM structure of SARS-CoV-2 Spike glycoprotein (one RBD up) in complex with COVOX-159 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-159 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND6
 
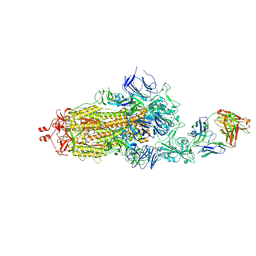 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-40 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
2VTP
 
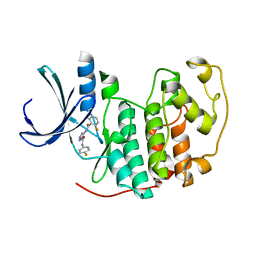 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, {[(2,6-difluorophenyl)carbonyl]amino}-N-(4-fluorophenyl)-1H-pyrazole-3-carboxamide | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
7NDB
 
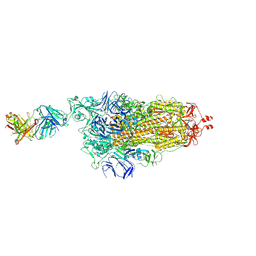 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-253H165L Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND8
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-384 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-384 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
8QV3
 
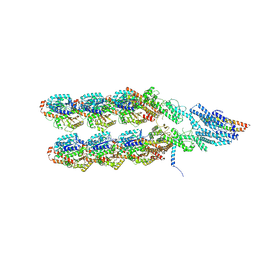 | | Structure of the y-Tubulin Small Complex (yTuSC) as part of the native y-Tubulin Ring Complex (yTuRC) capping microtubule minus ends at the spindle pole body | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Spindle pole body component, Spindle pole body component 110, ... | | Authors: | Dendooven, T, Yatskevich, S, Burt, A, Bellini, D, Kilmartin, J, Barford, D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structure of the native gamma-tubulin ring complex capping spindle microtubules.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4AFS
 
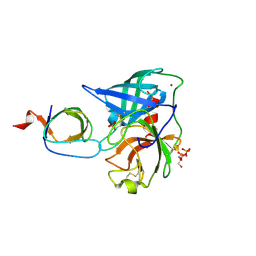 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, FYNOMER, GLYCEROL, ... | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
4AG2
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHYMASE, ... | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
7NDV
 
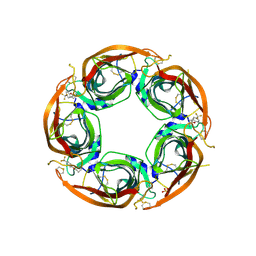 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001888. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[4-(trifluoromethyl)phenoxy]piperidine, Acetylcholine-binding protein, ... | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2021-02-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of fragments inducing conformational effects in dynamic proteins using a second-harmonic generation biosensor
RSC Advances, 11, 2021
|
|
8QV2
 
 | | Structure of the native y-Tubulin Ring Complex (yTuRC) capping microtubule minus ends at the spindle pole body | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Spindle pole body component, ... | | Authors: | Dendooven, T, Yatskevich, S, Burt, A, Bellini, D, Kilmartin, J, Barford, D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Structure of the native gamma-tubulin ring complex capping spindle microtubules.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7NDP
 
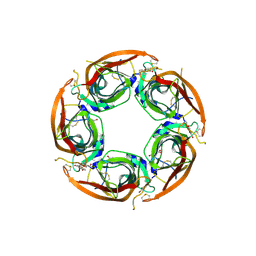 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001856. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-bromanylspiro[3~{H}-chromene-2,4'-piperidine]-4-one, ... | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2021-02-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of fragments inducing conformational effects in dynamic proteins using a second-harmonic generation biosensor.
Rsc Adv, 11, 2021
|
|
4AG1
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, FYNOMER, SULFATE ION | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
4AFU
 
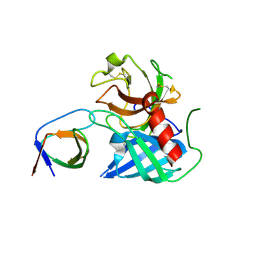 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, FYNOMER | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
3HUO
 
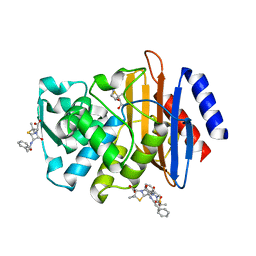 | | X-ray crystallographic structure of CTX-M-9 S70G in complex with benzylpenicillin | | Descriptor: | (2R,4S)-2-{(R)-carboxy[(phenylacetyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CTX-M-9 extended-spectrum beta-lactamase, PENICILLIN G | | Authors: | Delmas, J, Leyssene, D, Dubois, D, Robin, F, Bonnet, R. | | Deposit date: | 2009-06-15 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dynamic view of the early and late steps of the catalytic mechanism mediated by the emerging enzymes CTX-M.
To be Published
|
|
1CGF
 
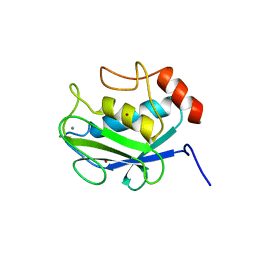 | | CRYSTAL STRUCTURES OF RECOMBINANT 19-KDA HUMAN FIBROBLAST COLLAGENASE COMPLEXED TO ITSELF | | Descriptor: | CALCIUM ION, FIBROBLAST COLLAGENASE, ZINC ION | | Authors: | Lovejoy, B, Hassell, A.M, Luther, M.A, Weigl, D, Jordan, S.R. | | Deposit date: | 1994-02-03 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of recombinant 19-kDa human fibroblast collagenase complexed to itself.
Biochemistry, 33, 1994
|
|
6DLM
 
 | | DHD127 | | Descriptor: | DHD127_A, DHD127_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-01 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
5NM2
 
 | | A2A Adenosine receptor cryo structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Dore, A.S, Geng, T, Cooke, R, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
3IPU
 
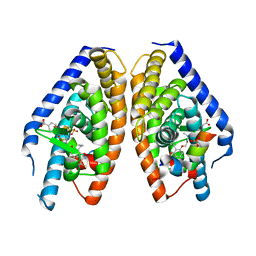 | | X-ray structure of benzisoxazole urea synthetic agonist bound to the LXR-alpha | | Descriptor: | 4-{[methyl(3-{[7-propyl-3-(trifluoromethyl)-1,2-benzisoxazol-6-yl]oxy}propyl)carbamoyl]amino}benzoic acid, Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha, ... | | Authors: | Fradera, X, Vu, D, Nimz, O, Skene, R, Hosfield, D, Wijnands, R, Cooke, A.J, Haunso, A, King, A, Bennet, D.J, McGuire, R, Uitdehaag, J.C.M. | | Deposit date: | 2009-08-18 | | Release date: | 2010-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of the LXRalpha LBD in its homodimeric form and implications for heterodimer signaling.
J.Mol.Biol., 399, 2010
|
|
4QR1
 
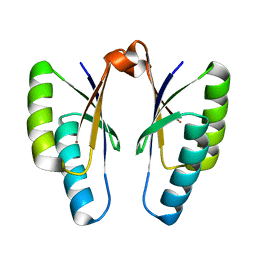 | | Crystal structure of Streptococcus pyogenes Cas2 at pH 6.5 | | Descriptor: | CRISPR-associated endoribonuclease Cas2 | | Authors: | Bae, E, Ka, D, Kim, D. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Structural and functional characterization of Streptococcus pyogenes Cas2 protein under different pH conditions
Biochem.Biophys.Res.Commun., 451, 2014
|
|
6DLC
 
 | |
6RC9
 
 | |
6QG9
 
 | | Crystal structure of Ideonella sakaiensis MHETase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
