4KMR
 
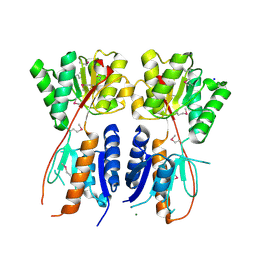 | | Structure of a putative transcriptional regulator of LacI family from Sanguibacter keddieii DSM 10542. | | Descriptor: | MAGNESIUM ION, SODIUM ION, Transcriptional regulator, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-08 | | Release date: | 2013-06-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a putative transcriptional regulator of LacI family from Sanguibacter keddieii DSM 10542.
To be Published
|
|
3U2E
 
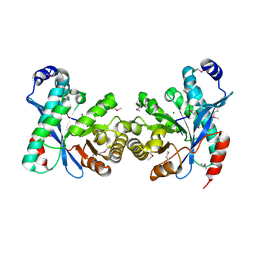 | | EAL domain of phosphodiesterase PdeA in complex with 5'-pGpG and Mg++ | | Descriptor: | GGDEF family protein, MAGNESIUM ION, RNA (5'-R(P*GP*G)-3') | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Massa, C, Schirmer, T, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-10-03 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | EAL domain from Caulobacter crescentus in complex with 5'-pGpG and Mg++
To be Published
|
|
4MPY
 
 | | 1.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus (IDP00699) in complex with NAD+ | | Descriptor: | Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-14 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based mutational studies of substrate inhibition of betaine aldehyde dehydrogenase BetB from Staphylococcus aureus.
Appl.Environ.Microbiol., 80, 2014
|
|
4MPB
 
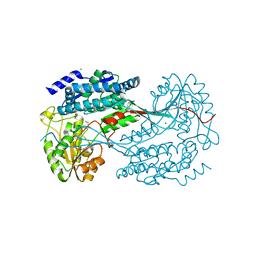 | | 1.7 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus | | Descriptor: | Betaine aldehyde dehydrogenase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based mutational studies of substrate inhibition of betaine aldehyde dehydrogenase BetB from Staphylococcus aureus.
Appl.Environ.Microbiol., 80, 2014
|
|
3FZY
 
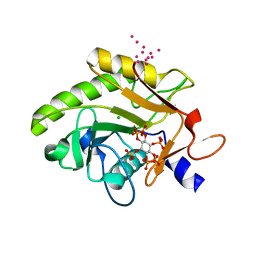 | | Crystal Structure of Pre-cleavage Form of Cysteine Protease Domain from Vibrio cholerae RtxA Toxin | | Descriptor: | CHLORIDE ION, INOSITOL HEXAKISPHOSPHATE, RTX toxin RtxA, ... | | Authors: | Shuvalova, L, Minasov, G, Prochazkova, K, Satchell, K.J.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-26 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and molecular mechanism for autoprocessing of MARTX toxin of Vibrio cholerae at multiple sites
J.Biol.Chem., 284, 2009
|
|
1RXQ
 
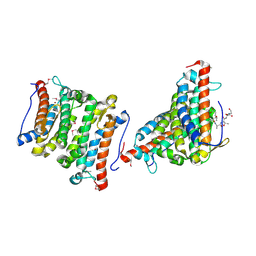 | | YfiT from Bacillus subtilis is a probable metal-dependent hydrolase with an unusual four-helix bundle topology | | Descriptor: | ALANINE, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Rajan, S.S, Yang, X, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-12-18 | | Release date: | 2004-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | YfiT from Bacillus subtilis Is a Probable Metal-Dependent Hydrolase with an Unusual Four-Helix Bundle Topology
Biochemistry, 43, 2004
|
|
5BXI
 
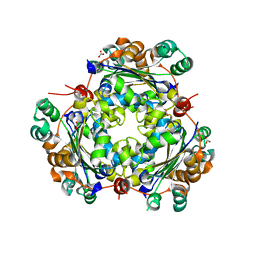 | | 1.7 Angstrom Resolution Crystal Structure of Putative Nucleoside Diphosphate Kinase from Toxoplasma gondii with Tyrosine of Tag Bound to Active Site | | Descriptor: | BICARBONATE ION, DI(HYDROXYETHYL)ETHER, Nucleoside diphosphate kinase | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-06-09 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
1QTX
 
 | |
1QS7
 
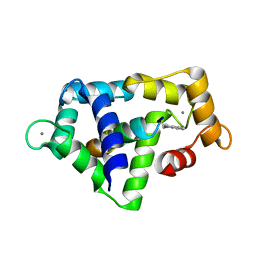 | |
2W27
 
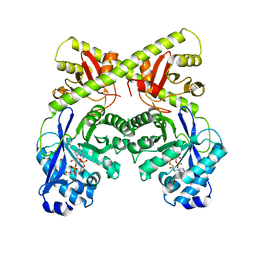 | | CRYSTAL STRUCTURE OF THE BACILLUS SUBTILIS YKUI PROTEIN, WITH AN EAL DOMAIN, IN COMPLEX WITH SUBSTRATE C-DI-GMP AND CALCIUM | | Descriptor: | CALCIUM ION, GUANOSINE-5'-MONOPHOSPHATE, YKUI PROTEIN | | Authors: | Padavattan, S, AndERSON, W.F, Schirmer, T. | | Deposit date: | 2008-10-24 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Ykui and its Complex with Second Messenger C-Di-Gmp Suggests Catalytic Mechanism of Phosphodiester Bond Cleavage by Eal Domains.
J.Biol.Chem., 284, 2009
|
|
7N1M
 
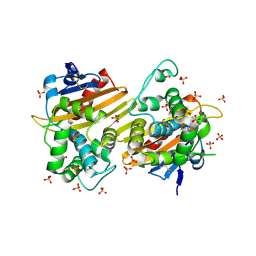 | | Crystal Structure of the Class D Beta-lactamase OXA-935 from Pseudomonas aeruginosa, Orthorhombic Crystal Form | | Descriptor: | Beta-lactamase OXA-935, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.B, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-27 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Functional and Structural Characterization of OXA-935, a Novel OXA-10-Family beta-Lactamase from Pseudomonas aeruginosa.
Antimicrob.Agents Chemother., 66, 2022
|
|
7L5R
 
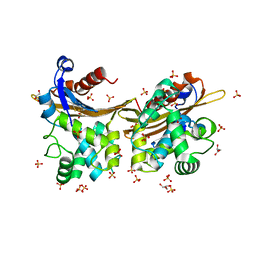 | | Crystal Structure of the Oxacillin-hydrolyzing Class D Extended-spectrum Beta-lactamase OXA-14 from Pseudomonas aeruginosa | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-22 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional and Structural Characterization of OXA-935, a Novel OXA-10-Family beta-Lactamase from Pseudomonas aeruginosa.
Antimicrob.Agents Chemother., 66, 2022
|
|
7L5V
 
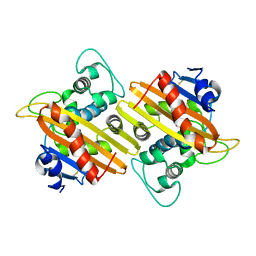 | | Crystal Structure of the Class D Beta-lactamase OXA-935 from Pseudomonas aeruginosa, Monoclinic Crystal Form | | Descriptor: | Beta-lactamase | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Functional and Structural Characterization of OXA-935, a Novel OXA-10-Family beta-Lactamase from Pseudomonas aeruginosa.
Antimicrob.Agents Chemother., 66, 2022
|
|
5I1T
 
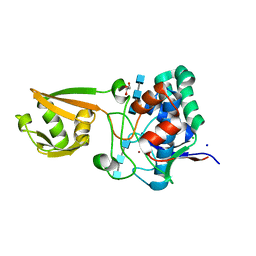 | | 2.6 Angstrom Resolution Crystal Structure of Stage II Sporulation Protein D (SpoIID) from Clostridium difficile in Complex with Triacetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nocadello, S, Minasov, G, Shuvalova, L, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-05 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the SpoIID Lytic Transglycosylases Essential for Bacterial Sporulation.
J.Biol.Chem., 291, 2016
|
|
5I82
 
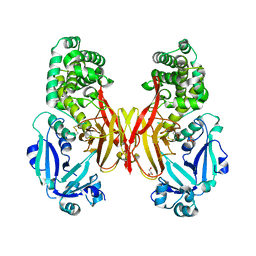 | | First Crystal Structure of E.coli Based Recombinant Diphtheria Toxin Mutant CRM197 | | Descriptor: | Diphtheria toxin, GLYCEROL, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Mishra, R.P.N, Goel, A, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-18 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and immunological characterization of E. coli derived recombinant CRM 197 protein used as carrier in conjugate vaccines.
Biosci.Rep., 38, 2018
|
|
7TL5
 
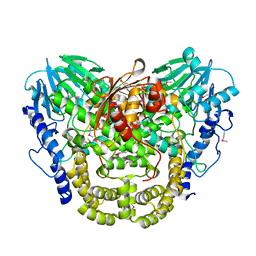 | | Crystal structure of putative hydrolase yjcS from Klebsiella pneumoniae. | | Descriptor: | 1,2-ETHANEDIOL, Lactamase_B domain-containing protein | | Authors: | Chang, C, Endres, M, Wu, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-18 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
3UPB
 
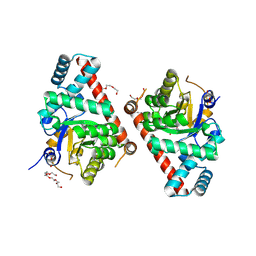 | | 1.5 Angstrom Resolution Crystal Structure of Transaldolase from Francisella tularensis in Covalent Complex with Arabinose-5-Phosphate | | Descriptor: | ARABINOSE-5-PHOSPHATE, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Light, S.H, Minasov, G, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-17 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Arabinose 5-phosphate covalently inhibits transaldolase.
J.Struct.Funct.Genom., 15, 2014
|
|
4RWR
 
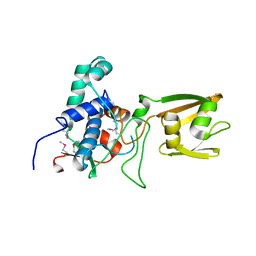 | | 2.1 Angstrom Crystal Structure of Stage II Sporulation Protein D from Bacillus anthracis | | Descriptor: | Stage II sporulation protein D | | Authors: | Minasov, G, Wawrzak, Z, Nocadello, S, Shuvalova, L, Dubrovska, I, Flores, K, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-05 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the SpoIID Lytic Transglycosylases Essential for Bacterial Sporulation.
J.Biol.Chem., 291, 2016
|
|
5T1V
 
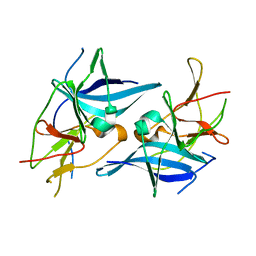 | | Crystal structure of Zika virus NS2B-NS3 protease in apo-form. | | Descriptor: | NS2B-NS3 protease,NS2B-NS3 protease | | Authors: | Nocadello, S, Light, S.H, Minasov, G, Shuvalova, L, Cardona-Correa, A.A, Ojeda, I, Vargas, J, Johnson, M.E, Lee, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-22 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of Zika virus NS2B-NS3 protease in apo-form.
To Be Published
|
|
6W61
 
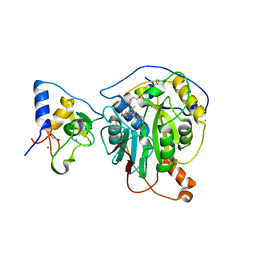 | | Crystal Structure of the methyltransferase-stimulatory factor complex of NSP16 and NSP10 from SARS CoV-2. | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, CHLORIDE ION, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine
Biorxiv, 2020
|
|
4NML
 
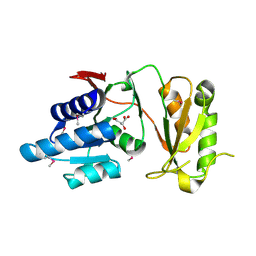 | | 2.60 Angstrom resolution crystal structure of putative ribose 5-phosphate isomerase from Toxoplasma gondii ME49 in complex with DL-Malic acid | | Descriptor: | CHLORIDE ION, D-MALATE, Ribulose 5-phosphate isomerase | | Authors: | Halavaty, A.S, Dubrovska, I, Flores, K, Shanmugam, D, Shuvalova, L, Roos, D, Ruan, J, Ngo, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4NOG
 
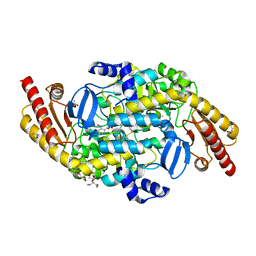 | | Crystal structure of a putative ornithine aminotransferase from Toxoplasma gondii ME49 in complex with pyrodoxal-5'-phosphate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Halavaty, A, Ruan, J, Shuvalova, L, Flores, K, Dubrovska, I, Ngo, H, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4NU7
 
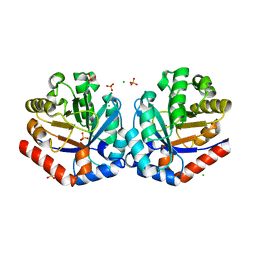 | | 2.05 Angstrom Crystal Structure of Ribulose-phosphate 3-epimerase from Toxoplasma gondii. | | Descriptor: | CHLORIDE ION, Ribulose-phosphate 3-epimerase, SULFATE ION, ... | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-03 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4O0N
 
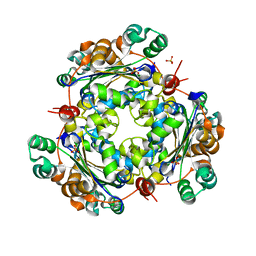 | | 2.4 Angstrom Resolution Crystal Structure of Putative Nucleoside Diphosphate Kinase from Toxoplasma gondii. | | Descriptor: | Nucleoside diphosphate kinase, SULFATE ION | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-13 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
