8Z9Q
 
 | | Cryo-EM structure of Thogoto virus polymerase in a replication reception conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(ATP))-R(P*GP*CP*AP*AP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z8X
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription initiation conformation | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z85
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 1 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-21 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
7DTD
 
 | | Voltage-gated sodium channel Nav1.1 and beta4 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 1 subunit alpha, ... | | Authors: | Yan, N, Pan, X, Li, Z, Huang, G. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Comparative structural analysis of human Na v 1.1 and Na v 1.5 reveals mutational hotspots for sodium channelopathies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8ZET
 
 | | Tp-PSI-FCPI-S in Thalassiosira pseudonana | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | Authors: | Feng, Y, Li, Z, Shen, J.R, Wang, W. | | Deposit date: | 2024-05-06 | | Release date: | 2024-12-25 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of PSI-FCPI from Thalassiosira pseudonana grown under high light provide evidence for convergent evolution and light-adaptive strategies in diatom FCPIs.
J Integr Plant Biol, 67, 2025
|
|
8YQF
 
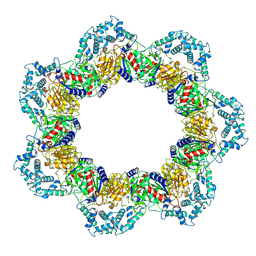 | |
8ZC5
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with D1F6 Fab, focused refinement of RBD region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC0
 
 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with 3 D1F6 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC2
 
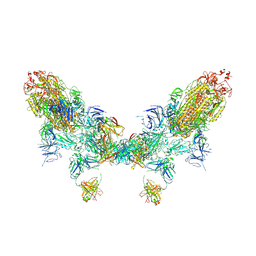 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with D1F6 Fab, head-to-head aggregate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.82 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZBZ
 
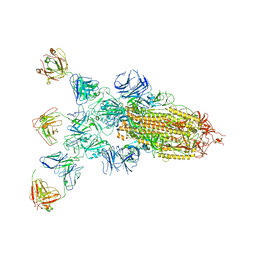 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with 3 D1F6 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.71 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC3
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with 3 D1F6 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC4
 
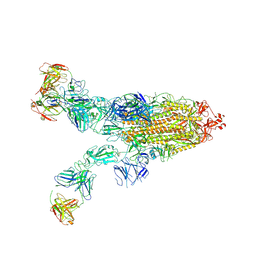 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with 3 D1F6 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZBY
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (x2-4P) in complex with 3 D1F6 Fabs (0 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC6
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with D1F6 Fab, head-to-head aggregate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.85 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC1
 
 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with D1F6 Fab, focused refinement of RBD region | | Descriptor: | Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, Spike protein S1 | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
6J8E
 
 | | Human Nav1.2-beta2-KIIIA ternary complex | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, X, Li, Z, Huang, X, Huang, G, Yan, N. | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for pore blockade of human Na+channel Nav1.2 by the mu-conotoxin KIIIA.
Science, 363, 2019
|
|
9ASL
 
 | |
6JBU
 
 | |
9ASK
 
 | |
9ASJ
 
 | |
6KEE
 
 | | Crystal structure of BRD4 Bromodomain1 with an inhibitor | | Descriptor: | 6-[2-[2,4-bis(fluoranyl)phenoxy]-5-(methylsulfonylmethyl)pyridin-3-yl]-8-methyl-2H-pyrrolo[1,2-a]pyrazin-1-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12154651 Å) | | Cite: | Crystal structure of BRD4 Bromodomain1 with an inhibitor
To be published
|
|
6KL9
 
 | | Structure of LbCas12a-crRNA complex bound to AcrVA4 (form A complex) | | Descriptor: | AcrVA4, LbCas12a, MAGNESIUM ION, ... | | Authors: | Peng, R, Li, Z, Xu, Y, He, S, Peng, Q, Shi, Y, Gao, G.F. | | Deposit date: | 2019-07-30 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insight into multistage inhibition of CRISPR-Cas12a by AcrVA4.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KED
 
 | | BRD4 Bromodomain1 with an inhibitor | | Descriptor: | 6-[2-[2,4-bis(fluoranyl)phenoxy]-5-(methylsulfonylmethyl)pyridin-3-yl]-8-methyl-2H-pyrrolo[1,2-d][1,2,4]triazin-1-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.548447 Å) | | Cite: | BRD4 Bromodomain1 with an inhibitor
To Be Published
|
|
6KLB
 
 | | Structure of LbCas12a-crRNA complex bound to AcrVA4 (form B complex) | | Descriptor: | AcrVA4, LbCas12a, MAGNESIUM ION, ... | | Authors: | Peng, R, Li, Z, Xu, Y, He, S, Peng, Q, Shi, Y, Gao, G.F. | | Deposit date: | 2019-07-30 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into multistage inhibition of CRISPR-Cas12a by AcrVA4.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KEG
 
 | | BRD4 Bromodomain1 with an inhibitor | | Descriptor: | 6-[2-[2,4-bis(fluoranyl)phenoxy]-5-(ethylsulfonylmethyl)pyridin-3-yl]-8-methyl-4H-pyrrolo[1,2-a]pyrazin-1-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.232 Å) | | Cite: | BRD4 Bromodomain1 with an inhibitor
To Be Published
|
|
