8FIW
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor Jun10221 | | Descriptor: | 3C-like proteinase nsp5, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]prop-2-enamide, N-([1,1'-biphenyl]-4-yl)-N-[(1S)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]prop-2-enamide | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2022-12-16 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Exploring diverse reactive warheads for the design of SARS-CoV-2 main protease inhibitors.
Eur.J.Med.Chem., 259, 2023
|
|
3LXG
 
 | | Crystal structure of rat phosphodiesterase 10A in complex with ligand WEB-3 | | Descriptor: | 2-methoxy-6,7-dimethyl-9-propylimidazo[1,5-a]pyrido[3,2-e]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Mosbacher, T, Jestel, A, Steinbacher, S. | | Deposit date: | 2010-02-25 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of imidazo[1,5-a]pyrido[3,2-e]pyrazines as a new class of phosphodiesterase 10A inhibitiors.
J.Med.Chem., 53, 2010
|
|
7DDD
 
 | | SARS-Cov2 S protein at close state | | Descriptor: | Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2020-10-28 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections.
Nat Commun, 12, 2021
|
|
7DK7
 
 | |
7DCX
 
 | |
7DK6
 
 | |
7DD8
 
 | |
7DK5
 
 | |
7DCC
 
 | |
7RN0
 
 | |
7RN1
 
 | |
6WNY
 
 | | Crystal structure of BACE1 in complex with (Z)-fluoro-olefin containing compound 15 | | Descriptor: | 6-[(Z)-2-{3-[(1S,5S,6S)-3-amino-5-methyl-1-(morpholine-4-carbonyl)-2-thia-4-azabicyclo[4.1.0]hept-3-en-5-yl]-4-fluorophenyl}-1-fluoroethenyl]pyridine-3-carbonitrile, Beta-secretase 1, IODIDE ION | | Authors: | Whittington, D.A. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The development of a structurally distinct series of BACE1 inhibitors via the (Z)-fluoro-olefin amide bioisosteric replacement.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7JM5
 
 | | Crystal structure of KDM4B in complex with QC6352 | | Descriptor: | 3-[({(1R)-6-[methyl(phenyl)amino]-1,2,3,4-tetrahydronaphthalen-1-yl}methyl)amino]pyridine-4-carboxylic acid, Lysine-specific demethylase 4B, NICKEL (II) ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-07-31 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting KDM4 for treating PAX3-FOXO1-driven alveolar rhabdomyosarcoma.
Sci Transl Med, 14, 2022
|
|
5TBM
 
 | | Crystal structure of PT2385 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 3-{[(1S)-2,2-difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1H-inden-4-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2016-09-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Small-Molecule Antagonist of HIF2 alpha Is Efficacious in Preclinical Models of Renal Cell Carcinoma.
Cancer Res., 76, 2016
|
|
3P17
 
 | | Thrombin Inhibition by Pyridin Derivatives | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-(pyridin-3-ylmethyl)-L-prolinamide, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2010-09-30 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Impact of ligand and protein desolvation on ligand binding to the S1 pocket of thrombin.
J.Mol.Biol., 418, 2012
|
|
3HC9
 
 | | Ferric Horse Heart Myoglobin; H64V mutant | | Descriptor: | Myoglobin, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yi, J, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2009-05-05 | | Release date: | 2009-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The distal pocket histidine residue in horse heart myoglobin directs the o-binding mode of nitrite to the heme iron.
J.Am.Chem.Soc., 131, 2009
|
|
3QTV
 
 | | Thrombin Inhibition by Pyridin Derivatives | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(1-methylpyridinium-4-yl)methyl]-L-prolinamide, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-02-23 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Impact of ligand and protein desolvation on ligand binding to the S1 pocket of thrombin
J.Mol.Biol., 418, 2012
|
|
3QWC
 
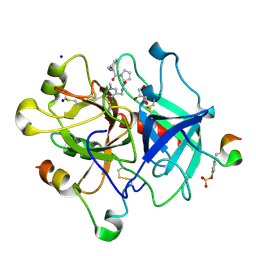 | | Thrombin Inhibition by Pyridin Derivatives | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(4-chloro-1-methylpyridinium-3-yl)methyl]-L-prolinamide, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Impact of ligand and protein desolvation on ligand binding to the S1 pocket of thrombin
J.Mol.Biol., 418, 2012
|
|
3HEN
 
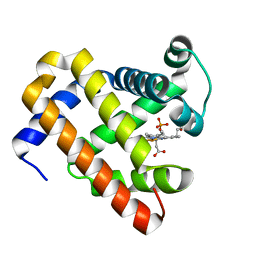 | | Ferric Horse Heart Myoglobin; H64V/V67R Mutant | | Descriptor: | Myoglobin, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yi, J, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2009-05-09 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The distal pocket histidine residue in horse heart myoglobin directs the o-binding mode of nitrite to the heme iron.
J.Am.Chem.Soc., 131, 2009
|
|
3HEO
 
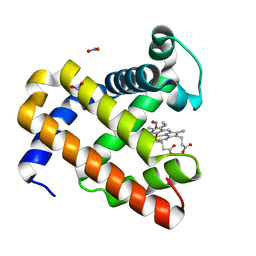 | | Ferric Horse Heart Myoglobin; H64V/V67R mutant, Nitrite Modified | | Descriptor: | Myoglobin, NITRITE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yi, J, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2009-05-09 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The distal pocket histidine residue in horse heart myoglobin directs the o-binding mode of nitrite to the heme iron.
J.Am.Chem.Soc., 131, 2009
|
|
3HEP
 
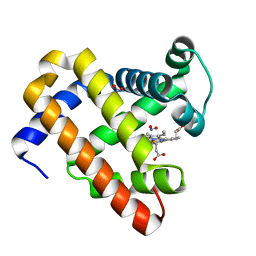 | | Ferric Horse Heart Myoglobin; H64V Mutant, Nitrite Modified | | Descriptor: | Myoglobin, NITRITE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yi, J, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2009-05-09 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The distal pocket histidine residue in horse heart myoglobin directs the o-binding mode of nitrite to the heme iron.
J.Am.Chem.Soc., 131, 2009
|
|
8WGW
 
 | | Local refinement of RBD-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
8WGV
 
 | | BA.2(S375) Spike (S6P)/hACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
7KX5
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with noncovalent inhibitor Jun8-76-3A | | Descriptor: | 3C-like proteinase, GLYCEROL, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]furan-2-carboxamide | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Di- and Trihaloacetamides as Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity.
J.Am.Chem.Soc., 143, 2021
|
|
3QTO
 
 | | Thrombin Inhibition by Pyridin Derivatives | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(1-methylpyridinium-3-yl)methyl]-L-prolinamide, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-02-23 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Impact of ligand and protein desolvation on ligand binding to the S1 pocket of thrombin
J.Mol.Biol., 418, 2012
|
|
