8QI7
 
 | |
8Q7S
 
 | | Crystal structure of the SARS-CoV-2 RBD (Wuhan) with neutralizing VHHs Ma6F06 and Re21H01 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Guttler, T, Aksu, M, Gorlich, D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8QU4
 
 | | NF-YB/C Heterodimer in Complex with a 13-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker in an alternative binding pose | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nuclear transcription factor Y subunit alpha, ... | | Authors: | Arbore, F, Durukan, C, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
8QU2
 
 | | NF-YB/C Heterodimer in Complex with a 16-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Durukan, C, Arbore, F, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
8QU3
 
 | | NF-YB/C Heterodimer in Complex with a 13-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, Nuclear transcription factor Y subunit alpha, ... | | Authors: | Arbore, F, Durukan, C, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
8RZA
 
 | | Ribonuclease W | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Probable ribonuclease FAU-1, ... | | Authors: | Vayssieres, M, Blaud, M, Leulliot, N. | | Deposit date: | 2024-02-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | RNase W, a conserved ribonuclease family with a novel active site.
Nucleic Acids Res., 2024
|
|
8R61
 
 | |
8RZE
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 10 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-pyridin-3-yl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RZC
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 11 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-imidazol-1-yl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RZD
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 9 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-(3-hydroxyphenyl)benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8QY4
 
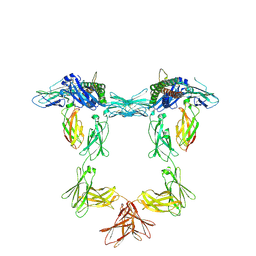 | | Structure of interleukin 11 (gp130 P496L mutant). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-11, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural insights into IL-11-mediated signalling and human IL6ST variant-associated immunodeficiency.
Nat Commun, 15, 2024
|
|
8QY5
 
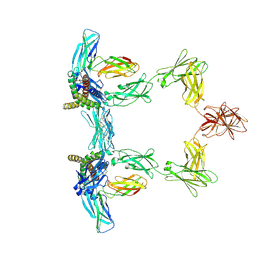 | | Structure of interleukin 6. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into IL-11-mediated signalling and human IL6ST variant-associated immunodeficiency.
Nat Commun, 15, 2024
|
|
8QY6
 
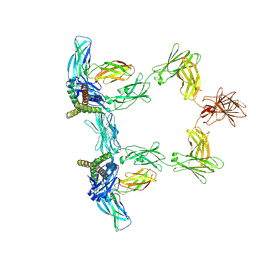 | | Structure of interleukin 6 (gp130 P496L mutant). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insights into IL-11-mediated signalling and human IL6ST variant-associated immunodeficiency.
Nat Commun, 15, 2024
|
|
8RIV
 
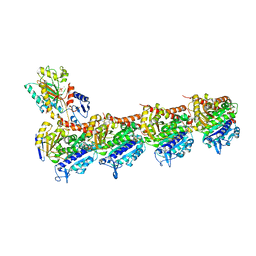 | | T2R-TTL-1-K08 complex | | Descriptor: | (4-fluoranyl-2-methyl-1~{H}-indol-5-yl) 3,4,5-trimethoxybenzenesulfonate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Boiarska, Z, Homer, J.A, Steinmetz, M.O, Moses, J.E, Prota, A.E.P. | | Deposit date: | 2023-12-19 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Modular synthesis of functional libraries by accelerated SuFEx click chemistry.
Chem Sci, 15, 2024
|
|
8RIW
 
 | | T2R-TTL-1-L01 complex | | Descriptor: | (2-methyl-1~{H}-indol-5-yl) 3,4,5-trimethoxybenzenesulfonate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E.P, Boiarska, Z, Homer, J.A, Steinmetz, M.O, Moses, J.E. | | Deposit date: | 2023-12-19 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Modular synthesis of functional libraries by accelerated SuFEx click chemistry.
Chem Sci, 15, 2024
|
|
8RUU
 
 | | Fabs derived from bimekizumab in complex with IL-17F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoblobulin heavy chain, Immunoblobulin light chain, ... | | Authors: | Adams, R, Lawson, A.D.G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of Bimekizumab Fab Fragment in Complex with IL-17F Provides Molecular Basis for Dual IL-17A and IL-17F Inhibition.
J Invest Dermatol., 144, 2024
|
|
8QZO
 
 | |
8ROO
 
 | | Crystal structure of HLA B*18:01 in complex with YERMCNIL, an 8-mer epitope from Influenza A | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Influenza A derived peptide/Nucleoprotein, ... | | Authors: | Murdolo, L.D, Maddumage, J.C, Gras, S. | | Deposit date: | 2024-01-11 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterisation of novel influenza-derived HLA-B*18:01-restricted epitopes.
Clin Transl Immunology, 13, 2024
|
|
8ROP
 
 | | Crystal structure of HLA B*18:01 in complex with QEIRTFSF, an 8-mer epitope from Influenza A | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Murdolo, L.D, Maddumage, J.C, Gras, S. | | Deposit date: | 2024-01-12 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Characterisation of novel influenza-derived HLA-B*18:01-restricted epitopes.
Clin Transl Immunology, 13, 2024
|
|
7JY2
 
 | | Z-DNA joint X-ray/Neutron | | Descriptor: | Chains: A,B | | Authors: | Harp, J.M, Coates, L, Egli, M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | Water structure around a left-handed Z-DNA fragment analyzed by cryo neutron crystallography.
Nucleic Acids Res., 49, 2021
|
|
8ASO
 
 | | Nickel(II) bound to a non-canonical quadruplex | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3'), NICKEL (II) ION | | Authors: | Lambert, M.C, Hall, J.P. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Identifying metal-DNA binding sites, what is the best method to get transition metals into a crystal system?
To Be Published
|
|
8ASM
 
 | | Cobalt(II) bound to a non-canonical quadruplex | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3') | | Authors: | Lambert, M.C, Hall, J.P. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identifying metal-DNA binding sites, what is the best method to get transition metals into a crystal system?
To Be Published
|
|
7JIZ
 
 | |
7KWO
 
 | | rFVIIIFc-VWF-XTEN (BIVV001) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Fuller, J.R, Batchelor, J.D. | | Deposit date: | 2020-12-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular determinants of the factor VIII/von Willebrand factor complex revealed by BIVV001 cryo-electron microscopy.
Blood, 137, 2021
|
|
7JMC
 
 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
